1AMW
 
 | |
4APE
 
 | | THE ACTIVE SITE OF ASPARTIC PROTEINASES | | 分子名称: | ENDOTHIAPEPSIN | | 著者 | Pearl, L.H, Sewell, B.T, Jenkins, J.A, Cooper, J.B, Blundell, T.L. | | 登録日 | 1986-06-09 | | 公開日 | 1986-07-14 | | 最終更新日 | 2019-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The Active Site of Aspartic Proteinases
FEBS Lett., 174, 1984
|
|
1AM1
 
 | |
1QNL
 
 | |
1GXR
 
 | | WD40 Region of Human Groucho/TLE1 | | 分子名称: | CALCIUM ION, TRANSDUCIN-LIKE ENHANCER PROTEIN 1 | | 著者 | Pearl, L.H, Roe, S.M, Pickles, L.M. | | 登録日 | 2002-04-10 | | 公開日 | 2002-06-13 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Crystal Structure of the C-Terminal Wd40 Repeat Domain of the Human Groucho/Tle1 Transcriptional Corepressor
Structure, 10, 2002
|
|
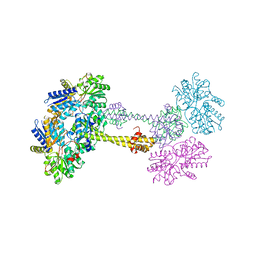
1QO0
 
 | |
1PEA
 
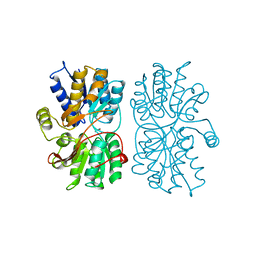 | |
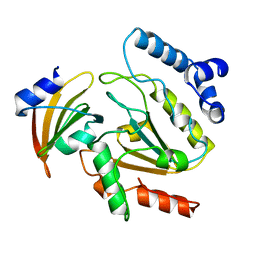
1UDI
 
 | |
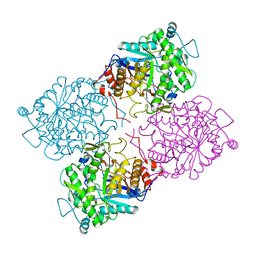
1GOW
 
 | |
1UDH
 
 | |
1UDG
 
 | |
1LAU
 
 | | URACIL-DNA GLYCOSYLASE | | 分子名称: | DNA (5'-D(*TP*TP*T)-3'), PROTEIN (URACIL-DNA GLYCOSYLASE (E.C.3.2.2.-)) | | 著者 | Pearl, L.H, Savva, R. | | 登録日 | 1996-01-03 | | 公開日 | 1996-06-10 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The structural basis of specific base-excision repair by uracil-DNA glycosylase.
Nature, 373, 1995
|
|
5K0Z
 
 | | Cryo-EM structure of lactate dehydrogenase (LDH) in inhibitor-bound state | | 分子名称: | L-lactate dehydrogenase B chain | | 著者 | Merk, A, Bartesaghi, A, Banerjee, S, Falconieri, V, Rao, P, Earl, L, Milne, J, Subramaniam, S. | | 登録日 | 2016-05-17 | | 公開日 | 2016-06-08 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Breaking Cryo-EM Resolution Barriers to Facilitate Drug Discovery.
Cell, 165, 2016
|
|
5K12
 
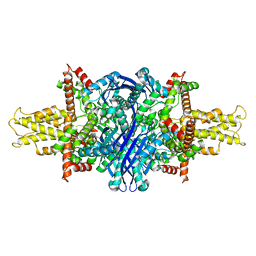 | | Cryo-EM structure of glutamate dehydrogenase at 1.8 A resolution | | 分子名称: | Glutamate dehydrogenase 1, mitochondrial | | 著者 | Merk, A, Bartesaghi, A, Banerjee, S, Falconieri, V, Rao, P, Earl, L, Milne, J, Subramaniam, S. | | 登録日 | 2016-05-17 | | 公開日 | 2016-06-08 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (1.8 Å) | | 主引用文献 | Breaking Cryo-EM Resolution Barriers to Facilitate Drug Discovery.
Cell, 165, 2016
|
|
5K10
 
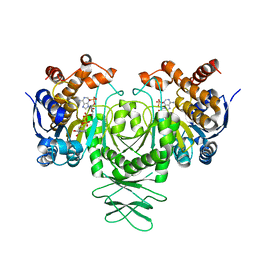 | | Cryo-EM structure of isocitrate dehydrogenase (IDH1) | | 分子名称: | Isocitrate dehydrogenase [NADP] cytoplasmic, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Merk, A, Bartesaghi, A, Banerjee, S, Falconieri, V, Rao, P, Earl, L, Milne, J, Subramaniam, S. | | 登録日 | 2016-05-17 | | 公開日 | 2016-06-08 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Breaking Cryo-EM Resolution Barriers to Facilitate Drug Discovery.
Cell, 165, 2016
|
|
5K11
 
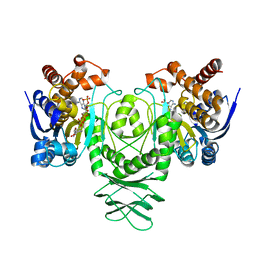 | | Cryo-EM structure of isocitrate dehydrogenase (IDH1) in inhibitor-bound state | | 分子名称: | Isocitrate dehydrogenase [NADP] cytoplasmic, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Merk, A, Bartesaghi, A, Banerjee, S, Falconieri, V, Rao, P, Earl, L, Milne, J, Subramaniam, S. | | 登録日 | 2016-05-17 | | 公開日 | 2016-06-08 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Breaking Cryo-EM Resolution Barriers to Facilitate Drug Discovery.
Cell, 165, 2016
|
|
3JD0
 
 | | Glutamate dehydrogenase in complex with GTP | | 分子名称: | GUANOSINE-5'-TRIPHOSPHATE, Glutamate dehydrogenase 1, mitochondrial | | 著者 | Borgnia, M.J, Banerjee, S, Merk, A, Matthies, D, Bartesaghi, A, Rao, P, Pierson, J, Earl, L.A, Falconieri, V, Subramaniam, S, Milne, J.L.S. | | 登録日 | 2016-03-28 | | 公開日 | 2016-04-27 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (3.47 Å) | | 主引用文献 | Using Cryo-EM to Map Small Ligands on Dynamic Metabolic Enzymes: Studies with Glutamate Dehydrogenase.
Mol.Pharmacol., 89, 2016
|
|
3JD4
 
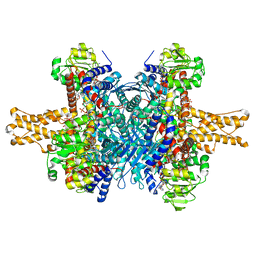 | | Glutamate dehydrogenase in complex with NADH and GTP, closed conformation | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GUANOSINE-5'-TRIPHOSPHATE, Glutamate dehydrogenase 1, ... | | 著者 | Borgnia, M.J, Banerjee, S, Merk, A, Matthies, D, Bartesaghi, A, Rao, P, Pierson, J, Earl, L.A, Falconieri, V, Subramaniam, S, Milne, J.L.S. | | 登録日 | 2016-03-28 | | 公開日 | 2016-04-27 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Using Cryo-EM to Map Small Ligands on Dynamic Metabolic Enzymes: Studies with Glutamate Dehydrogenase.
Mol.Pharmacol., 89, 2016
|
|
3JD1
 
 | | Glutamate dehydrogenase in complex with NADH, closed conformation | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Glutamate dehydrogenase 1, mitochondrial | | 著者 | Borgnia, M.J, Banerjee, S, Merk, A, Matthies, D, Bartesaghi, A, Rao, P, Pierson, J, Earl, L.A, Falconieri, V, Subramaniam, S, Milne, J.L.S. | | 登録日 | 2016-03-28 | | 公開日 | 2016-04-27 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Using Cryo-EM to Map Small Ligands on Dynamic Metabolic Enzymes: Studies with Glutamate Dehydrogenase.
Mol.Pharmacol., 89, 2016
|
|
3JD3
 
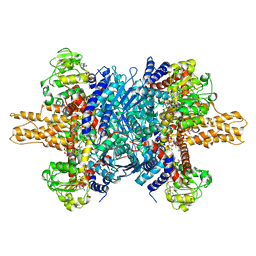 | | Glutamate dehydrogenase in complex with NADH and GTP, open conformation | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GUANOSINE-5'-TRIPHOSPHATE, Glutamate dehydrogenase 1, ... | | 著者 | Borgnia, M.J, Banerjee, S, Merk, A, Matthies, D, Bartesaghi, A, Rao, P, Pierson, J, Earl, L.A, Falconieri, V, Subramaniam, S, Milne, J.L.S. | | 登録日 | 2016-03-28 | | 公開日 | 2016-04-27 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Using Cryo-EM to Map Small Ligands on Dynamic Metabolic Enzymes: Studies with Glutamate Dehydrogenase.
Mol.Pharmacol., 89, 2016
|
|
6B44
 
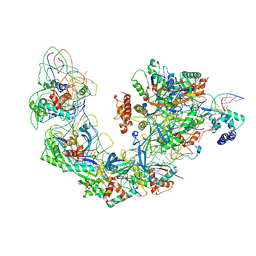 | | Cryo-EM structure of Type I-F CRISPR crRNA-guided Csy surveillance complex with bound target dsDNA | | 分子名称: | CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, CRISPR-associated protein Csy2, ... | | 著者 | Guo, T.W, Bartesaghi, A, Yang, H, Falconieri, V, Rao, P, Merk, A, Fox, T, Earl, L, Patel, D.J, Subramaniam, S. | | 登録日 | 2017-09-25 | | 公開日 | 2017-10-18 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Cryo-EM Structures Reveal Mechanism and Inhibition of DNA Targeting by a CRISPR-Cas Surveillance Complex.
Cell, 171, 2017
|
|
3JCZ
 
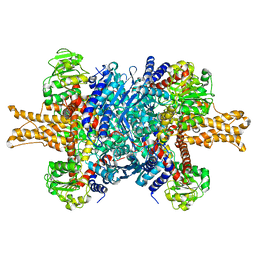 | | Structure of bovine glutamate dehydrogenase in the unliganded state | | 分子名称: | Glutamate dehydrogenase 1, mitochondrial | | 著者 | Borgnia, M.J, Banerjee, S, Merk, A, Matthies, D, Bartesaghi, A, Rao, P, Pierson, J, Earl, L.A, Falconieri, V, Subramaniam, S, Milne, J.L.S. | | 登録日 | 2016-03-27 | | 公開日 | 2016-04-27 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (3.26 Å) | | 主引用文献 | Using Cryo-EM to Map Small Ligands on Dynamic Metabolic Enzymes: Studies with Glutamate Dehydrogenase.
Mol.Pharmacol., 89, 2016
|
|
6B45
 
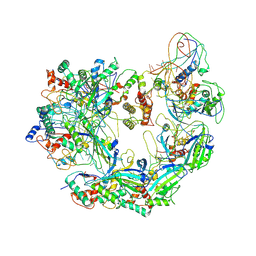 | | Cryo-EM structure of Type I-F CRISPR crRNA-guided Csy surveillance complex | | 分子名称: | CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, CRISPR-associated protein Csy2, ... | | 著者 | Guo, T.W, Bartesaghi, A, Yang, H, Falconieri, V, Rao, P, Merk, A, Fox, T, Earl, L, Patel, D.J, Subramaniam, S. | | 登録日 | 2017-09-25 | | 公開日 | 2017-10-18 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Cryo-EM Structures Reveal Mechanism and Inhibition of DNA Targeting by a CRISPR-Cas Surveillance Complex.
Cell, 171, 2017
|
|
6B47
 
 | | Cryo-EM structure of Type I-F CRISPR crRNA-guided Csy surveillance complex with bound anti-CRISPR protein AcrF2 | | 分子名称: | Anti-CRISPR protein AcrF2, CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, ... | | 著者 | Guo, T.W, Bartesaghi, A, Yang, H, Falconieri, V, Rao, P, Merk, A, Fox, T, Earl, L, Patel, D.J, Subramaniam, S. | | 登録日 | 2017-09-25 | | 公開日 | 2017-10-18 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Cryo-EM Structures Reveal Mechanism and Inhibition of DNA Targeting by a CRISPR-Cas Surveillance Complex.
Cell, 171, 2017
|
|
6B48
 
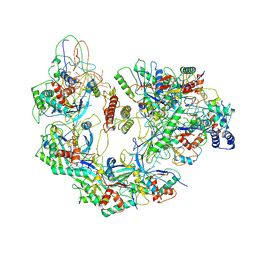 | | Cryo-EM structure of Type I-F CRISPR crRNA-guided Csy surveillance complex with bound anti-CRISPR protein AcrF10 | | 分子名称: | Anti-CRISPR protein AcrF10, CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, ... | | 著者 | Guo, T.W, Bartesaghi, A, Yang, H, Falconieri, V, Rao, P, Merk, A, Fox, T, Earl, L, Patel, D.J, Subramaniam, S. | | 登録日 | 2017-09-25 | | 公開日 | 2017-10-18 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Cryo-EM Structures Reveal Mechanism and Inhibition of DNA Targeting by a CRISPR-Cas Surveillance Complex.
Cell, 171, 2017
|
|
