5NN7
 
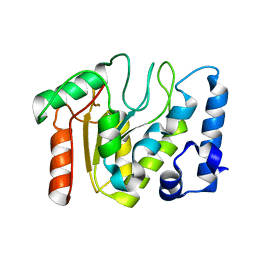 | | KSHV uracil-DNA glycosylase, apo form | | Descriptor: | Uracil-DNA glycosylase | | Authors: | Earl, C, Bagneris, C, Cole, A.R, Barrett, T, Savva, R. | | Deposit date: | 2017-04-08 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A structurally conserved motif in gamma-herpesvirus uracil-DNA glycosylases elicits duplex nucleotide-flipping.
Nucleic Acids Res., 46, 2018
|
|
5NNU
 
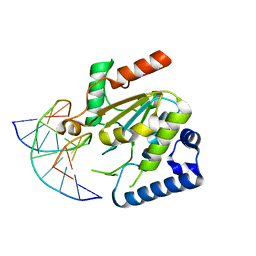 | | KSHV uracil-DNA glycosylase, product complex with dsDNA exhibiting duplex nucleotide flipping | | Descriptor: | DNA, DNA containing an abasic site, Uracil-DNA glycosylase | | Authors: | Earl, C, Bagneris, C, Barrett, T, Savva, R. | | Deposit date: | 2017-04-10 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | A structurally conserved motif in gamma-herpesvirus uracil-DNA glycosylases elicits duplex nucleotide-flipping.
Nucleic Acids Res., 46, 2018
|
|
5NNH
 
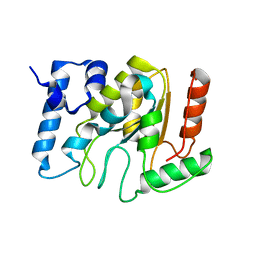 | | KSHV uracil-DNA glycosylase, apo form | | Descriptor: | SULFATE ION, Uracil-DNA glycosylase | | Authors: | Earl, C, Bagneris, C, Cole, A.R, Barrett, T, Savva, R. | | Deposit date: | 2017-04-09 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A structurally conserved motif in gamma-herpesvirus uracil-DNA glycosylases elicits duplex nucleotide-flipping.
Nucleic Acids Res., 46, 2018
|
|
7AB8
 
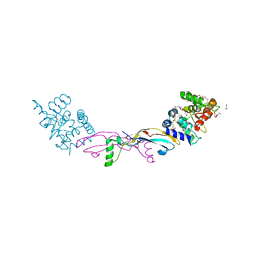 | | Crystal structure of a GDNF-GFRalpha1 complex | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Adams, S.E, Earl, C.P, Purkiss, A.G, McDonald, N.Q. | | Deposit date: | 2020-09-07 | | Release date: | 2021-01-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A two-site flexible clamp mechanism for RET-GDNF-GFR alpha 1 assembly reveals both conformational adaptation and strict geometric spacing.
Structure, 29, 2021
|
|
7AML
 
 | | RET/GDNF/GFRa1 extracellular complex Cryo-EM structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GDNF family receptor alpha, ... | | Authors: | Adams, S.E, Earl, C.P, Purkiss, A.G, McDonald, N.Q. | | Deposit date: | 2020-10-09 | | Release date: | 2021-01-13 | | Last modified: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | A two-site flexible clamp mechanism for RET-GDNF-GFR alpha 1 assembly reveals both conformational adaptation and strict geometric spacing.
Structure, 29, 2021
|
|
7R5Y
 
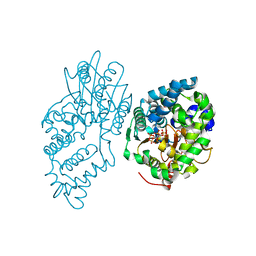 | |
7ZGF
 
 | |
7ZGH
 
 | |
7ZGG
 
 | |
6GIZ
 
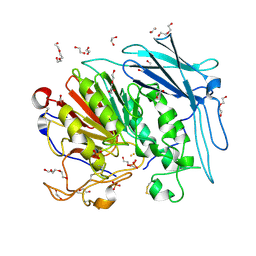 | | PURPLE ACID PHYTASE FROM WHEAT ISOFORM B2 - SUBSTRATE COMPLEX | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Faba-Rodriguez, R, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2018-05-15 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structure of a cereal purple acid phytase provides new insights to phytate degradation in plants.
Plant Commun., 3, 2022
|
|
7PUP
 
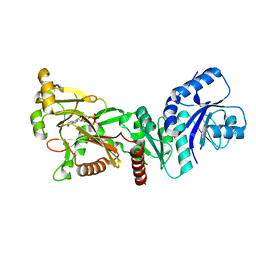 | | INOSITOL 1,3,4-TRISPHOSPHATE 5/6-KINASE 4 from Arabidopsis thaliana (AtITPK4) in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Inositol 1,3,4-trisphosphate 5/6-kinase 4 | | Authors: | Whitfield, H, He, S, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2021-09-30 | | Release date: | 2022-10-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Diversification in the inositol tris/tetrakisphosphate kinase (ITPK) family: crystal structure and enzymology of the outlier AtITPK4.
Biochem.J., 480, 2023
|
|
8OXE
 
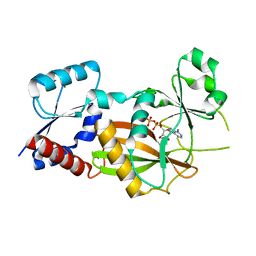 | | Inositol 1,3,4-trisphosphate 5/6-kinase 1 from Solanum tuberosum (StITPK1) in complex with ADP/Mg2+ | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Inositol-tetrakisphosphate 1-kinase, MAGNESIUM ION | | Authors: | Faba-Rodriguez, R, Li, A.W.H, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2023-05-02 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal Structure and Enzymology of Solanum tuberosum Inositol Tris/Tetrakisphosphate Kinase 1 ( St ITPK1).
Biochemistry, 63, 2024
|
|
6FL8
 
 | | Inositol 1,3,4,5,6-pentakisphosphate 2-kinase from A. thaliana in complex with purpurogallin and ADP | | Descriptor: | 1,2-ETHANEDIOL, 2,3,4,6-tetrahydroxy-5H-benzo[7]annulen-5-one, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Whitfield, H.L, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2018-01-25 | | Release date: | 2018-09-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Fluorescent Probe Identifies Active Site Ligands of Inositol Pentakisphosphate 2-Kinase.
J. Med. Chem., 61, 2018
|
|
6FL3
 
 | | Inositol 1,3,4,5,6-pentakisphosphate 2-kinase from A. thaliana in complex with myo-IP5 and ADP | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-DIPHOSPHATE, Inositol-pentakisphosphate 2-kinase, ... | | Authors: | Whitfield, H.L, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2018-01-25 | | Release date: | 2018-09-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | A Fluorescent Probe Identifies Active Site Ligands of Inositol Pentakisphosphate 2-Kinase.
J. Med. Chem., 61, 2018
|
|
6FJK
 
 | | Inositol 1,3,4,5,6-pentakisphosphate 2-kinase from A. thaliana in complex with myo-IP6 and ADP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-DIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Whitfield, H.L, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2018-01-22 | | Release date: | 2018-09-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.025 Å) | | Cite: | A Fluorescent Probe Identifies Active Site Ligands of Inositol Pentakisphosphate 2-Kinase.
J. Med. Chem., 61, 2018
|
|
6GJ2
 
 | | PURPLE ACID PHYTASE FROM WHEAT ISOFORM B2 - COMPLEX WITH INOSITOL HEXASULPHATE | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Faba-Rodriguez, R, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2018-05-15 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structure of a cereal purple acid phytase provides new insights to phytate degradation in plants.
Plant Commun., 3, 2022
|
|
6GJ9
 
 | |
6GIT
 
 | | PURPLE ACID PHYTASE FROM WHEAT ISOFORM B2 - PRODUCT COMPLEX | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Faba-Rodriguez, R, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2018-05-15 | | Release date: | 2019-11-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.418 Å) | | Cite: | Structure of a cereal purple acid phytase provides new insights to phytate degradation in plants.
Plant Commun., 3, 2022
|
|
6GFG
 
 | | Inositol 1,3,4,5,6-pentakisphosphate 2-kinase from A. thaliana in complex with D-chiro-IP6 and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, D-chiro inositol hexakisphosphate, Inositol-pentakisphosphate 2-kinase, ... | | Authors: | Whitfield, H.L, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2018-04-30 | | Release date: | 2018-09-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A Fluorescent Probe Identifies Active Site Ligands of Inositol Pentakisphosphate 2-Kinase.
J. Med. Chem., 61, 2018
|
|
6GJA
 
 | | PURPLE ACID PHYTASE FROM WHEAT ISOFORM B2 - H229A MUTANT | | Descriptor: | 1,2-ETHANEDIOL, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Faba-Rodriguez, R, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2018-05-16 | | Release date: | 2019-11-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of a cereal purple acid phytase provides new insights to phytate degradation in plants.
Plant Commun., 3, 2022
|
|
6GFH
 
 | | Inositol 1,3,4,5,6-pentakisphosphate 2-kinase from A. thaliana in complex with neo-IP5 and ATP | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Whitfield, H.L, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2018-04-30 | | Release date: | 2018-09-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A Fluorescent Probe Identifies Active Site Ligands of Inositol Pentakisphosphate 2-Kinase.
J. Med. Chem., 61, 2018
|
|
6RXG
 
 | |
6RXF
 
 | |
6RXD
 
 | |
6RXE
 
 | |
