2A8Y
 
 | | Crystal structure of 5'-deoxy-5'methylthioadenosine phosphorylase complexed with 5'-deoxy-5'methylthioadenosine and sulfate | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, 5'-methylthioadenosine phosphorylase (mtaP), SULFATE ION | | Authors: | Zhang, Y, Porcelli, M, Cacciapuoti, G, Ealick, S.E. | | Deposit date: | 2005-07-10 | | Release date: | 2006-03-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The crystal structure of 5'-deoxy-5'-methylthioadenosine phosphorylase II from Sulfolobus solfataricus, a thermophilic enzyme stabilized by intramolecular disulfide bonds.
J.Mol.Biol., 357, 2006
|
|
1DBT
 
 | | CRYSTAL STRUCTURE OF OROTIDINE 5'-MONOPHOSPHATE DECARBOXYLASE FROM BACILLUS SUBTILIS COMPLEXED WITH UMP | | Descriptor: | OROTIDINE 5'-PHOSPHATE DECARBOXYLASE, URIDINE-5'-MONOPHOSPHATE | | Authors: | Appleby, T.C, Kinsland, C.L, Begley, T.P, Ealick, S.E. | | Deposit date: | 1999-11-03 | | Release date: | 2000-03-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure and mechanism of orotidine 5'-monophosphate decarboxylase.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
2A9Z
 
 | | Crystal structure of T. gondii adenosine kinase complexed with N6-dimethyladenosine and AMP-PCP | | Descriptor: | 6N-DIMETHYLADENOSINE, CHLORIDE ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Zhang, Y, el Kouni, M.H, Ealick, S.E. | | Deposit date: | 2005-07-12 | | Release date: | 2006-07-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Substrate analogs induce an intermediate conformational change in Toxoplasma gondii adenosine kinase
Acta Crystallogr.,Sect.D, 63, 2007
|
|
1NG3
 
 | | Complex of ThiO (glycine oxidase) with acetyl-glycine | | Descriptor: | ACETYLAMINO-ACETIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, Glycine oxidase, ... | | Authors: | Settembre, E.C, Dorrestein, P.C, Park, J, Augustine, A, Begley, T.P, Ealick, S.E. | | Deposit date: | 2002-12-16 | | Release date: | 2003-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Mechanistic Studies on ThiO, a Glycine Oxidase Essential for Thiamin Biosynthesis in Bacillus subtilis
Biochemistry, 42, 2003
|
|
1NG4
 
 | | Structure of ThiO (glycine oxidase) from Bacillus subtilis | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Glycine oxidase, HYDROGEN PEROXIDE, ... | | Authors: | Settembre, E.C, Dorrestein, P.C, Park, J, Augustine, A, Begley, T.P, Ealick, S.E. | | Deposit date: | 2002-12-16 | | Release date: | 2003-04-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Mechanistic Studies on ThiO, a Glycine Oxidase Essential for Thiamin Biosynthesis in Bacillus subtilis
Biochemistry, 42, 2003
|
|
1OTP
 
 | | STRUCTURAL AND THEORETICAL STUDIES SUGGEST DOMAIN MOVEMENT PRODUCES AN ACTIVE CONFORMATION OF THYMIDINE PHOSPHORYLASE | | Descriptor: | THYMIDINE PHOSPHORYLASE | | Authors: | Pugmire, M.J, Cook, W.J, Jasanoff, A, Walter, M.R, Ealick, S.E. | | Deposit date: | 1997-11-09 | | Release date: | 1998-12-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and theoretical studies suggest domain movement produces an active conformation of thymidine phosphorylase.
J.Mol.Biol., 281, 1998
|
|
1N13
 
 | | The Crystal Structure of Pyruvoyl-dependent Arginine Decarboxylase from Methanococcus jannashii | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, AGMATINE, Pyruvoyl-dependent arginine decarboxylase alpha chain, ... | | Authors: | Tolbert, W.D, Graham, D.E, White, R.H, Ealick, S.E. | | Deposit date: | 2002-10-16 | | Release date: | 2003-03-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Pyruvoyl-Dependent Arginine Decarboxylase from Methanococcus jannaschii:
Crystal Structures of the Self-Cleaved and S53A Proenzyme Forms
Structure, 11, 2003
|
|
1S2D
 
 | | Purine 2'-Deoxyribosyl complex with arabinoside: Ribosylated Intermediate (AraA) | | Descriptor: | 2-deoxy-2-fluoro-alpha-D-arabinofuranose, ADENINE, Nucleoside 2-deoxyribosyltransferase | | Authors: | Anand, R, Kaminski, P.A, Ealick, S.E. | | Deposit date: | 2004-01-08 | | Release date: | 2004-03-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of purine 2'-deoxyribosyltransferase, substrate complexes, and the ribosylated enzyme intermediate at 2.0 A resolution.
Biochemistry, 43, 2004
|
|
1P9O
 
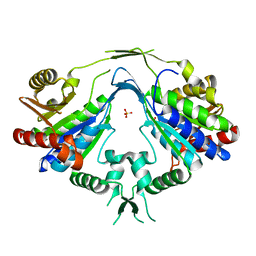 | | Crystal Structure of Phosphopantothenoylcysteine Synthetase | | Descriptor: | Phosphopantothenoylcysteine synthetase, SULFATE ION | | Authors: | Manoj, N, Strauss, E, Begley, T.P, Ealick, S.E. | | Deposit date: | 2003-05-12 | | Release date: | 2003-09-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of human phosphopantothenoylcysteine synthetase at 2.3 A resolution.
Structure, 11, 2003
|
|
1S2L
 
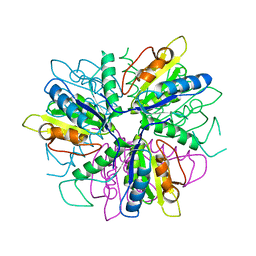 | |
1S2G
 
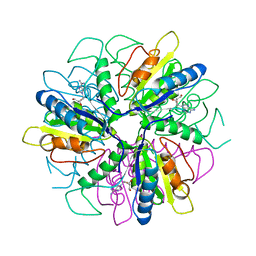 | | Purine 2'deoxyribosyltransferase + 2'-deoxyadenosine | | Descriptor: | (2R,3S,5R)-5-(6-amino-9H-purin-9-yl)-tetrahydro-2-(hydroxymethyl)furan-3-ol, purine trans deoxyribosylase | | Authors: | Anand, R, Kaminski, P.A, Ealick, S.E. | | Deposit date: | 2004-01-08 | | Release date: | 2004-03-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of purine 2'-deoxyribosyltransferase, substrate complexes, and the ribosylated enzyme intermediate at 2.0 A resolution.
Biochemistry, 43, 2004
|
|
1S2I
 
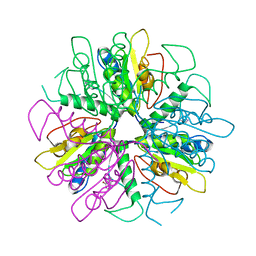 | | Purine 2'deoxyribosyltransferase + bromopurine | | Descriptor: | 6-BROMO-7H-PURINE, purine trans deoxyribosylase | | Authors: | Anand, R, Kaminski, P.A, Ealick, S.E. | | Deposit date: | 2004-01-08 | | Release date: | 2004-03-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structures of purine 2'-deoxyribosyltransferase, substrate complexes, and the ribosylated enzyme intermediate at 2.0 A resolution.
Biochemistry, 43, 2004
|
|
1S3F
 
 | | Purine 2'-deoxyribosyltransferase + selenoinosine | | Descriptor: | 9-(3,4-DIHYDROXY-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-2-YL)-1,9-DIHYDRO-PURINE-6-THIONE, purine trans deoxyribosylase | | Authors: | Anand, R, Kaminski, P.A, Ealick, S.E. | | Deposit date: | 2004-01-13 | | Release date: | 2004-03-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of purine 2'-deoxyribosyltransferase, substrate complexes, and the ribosylated enzyme intermediate at 2.0 A resolution.
Biochemistry, 43, 2004
|
|
1PK7
 
 | | Crystal Structure of E. coli purine nucleoside phosphorylase complexed with adenosine and sulfate/phosphate | | Descriptor: | ADENOSINE, PHOSPHATE ION, Purine nucleoside phosphorylase DeoD-type | | Authors: | Bennett, E.M, Li, C, Allan, P.W, Parker, W.B, Ealick, S.E. | | Deposit date: | 2003-06-05 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for substrate specificity of Escherichia coli purine nucleoside phosphorylase.
J.Biol.Chem., 278, 2003
|
|
1PK9
 
 | | Crystal Structure of E. coli purine nucleoside phosphorylase complexed with 2-fluoroadenosine and sulfate/phosphate | | Descriptor: | 2-(6-AMINO-2-FLUORO-PURIN-9-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, PHOSPHATE ION, Purine nucleoside phosphorylase DeoD-type | | Authors: | Bennett, E.M, Li, C, Allan, P.W, Parker, W.B, Ealick, S.E. | | Deposit date: | 2003-06-05 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for substrate specificity of Escherichia coli purine nucleoside phosphorylase.
J.Biol.Chem., 278, 2003
|
|
1PKE
 
 | | Crystal Structure of E. coli purine nucleoside phosphorylase complexed with 2-fluoro-2'-deoxyadenosine and sulfate/phosphate | | Descriptor: | 5-(6-AMINO-2-FLUORO-PURIN-9-YL)-2-HYDROXYMETHYL-TETRAHYDRO-FURAN-3-OL, PHOSPHATE ION, Purine nucleoside phosphorylase DeoD-type | | Authors: | Bennett, E.M, Li, C, Allan, P.W, Parker, W.B, Ealick, S.E. | | Deposit date: | 2003-06-05 | | Release date: | 2003-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for substrate specificity of Escherichia coli purine nucleoside phosphorylase.
J.Biol.Chem., 278, 2003
|
|
1PR4
 
 | | Escherichia coli Purine Nucleoside Phosphorylase Complexed with 9-beta-D-ribofuranosyl-6-methylthiopurine and Phosphate/Sulfate | | Descriptor: | 2-HYDROXYMETHYL-5-(6-METHYLSULFANYL-PURIN-9-YL)-TETRAHYDRO-FURAN-3,4-DIOL, PHOSPHATE ION, Purine nucleoside phosphorylase DeoD-type | | Authors: | Bennett, E.M, Li, C, Allan, P.W, Parker, W.B, Ealick, S.E. | | Deposit date: | 2003-06-19 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for substrate specificity of Escherichia coli purine nucleoside phosphorylase.
J.Biol.Chem., 278, 2003
|
|
1PW7
 
 | | Crystal Structure of E. coli purine nucleoside phosphorylase complexed with 9-beta-D-arabinofuranosyladenine and sulfate/phosphate | | Descriptor: | 2-(6-AMINO-PURIN-9-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, PHOSPHATE ION, Purine nucleoside phosphorylase DeoD-type | | Authors: | Bennett, E.M, Li, C, Allan, P.W, Parker, W.B, Ealick, S.E. | | Deposit date: | 2003-06-30 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for substrate specificity of Escherichia coli purine nucleoside phosphorylase.
J.Biol.Chem., 278, 2003
|
|
1PR1
 
 | | Escherichia coli Purine Nucleoside Phosphorylase Complexed with Formycin B and Phosphate/Sulfate | | Descriptor: | FORMYCIN B, PHOSPHATE ION, Purine nucleoside phosphorylase | | Authors: | Bennett, E.M, Li, C, Allan, P.W, Parker, W.B, Ealick, S.E. | | Deposit date: | 2003-06-19 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for substrate specificity of Escherichia coli purine nucleoside phosphorylase.
J.Biol.Chem., 278, 2003
|
|
1PR6
 
 | | Escherichia coli Purine Nucleoside Phosphorylase Complexed with 9-beta-D-xylofuranosyladenine and Phosphate/Sulfate | | Descriptor: | 2-(6-AMINO-OCTAHYDRO-PURIN-9-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, PHOSPHATE ION, Purine nucleoside phosphorylase DeoD-type | | Authors: | Bennett, E.M, Li, C, Allan, P.W, Parker, W.B, Ealick, S.E. | | Deposit date: | 2003-06-19 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for substrate specificity of Escherichia coli purine nucleoside phosphorylase.
J.Biol.Chem., 278, 2003
|
|
1N2M
 
 | | The S53A Proenzyme Structure of Methanococcus jannaschii. | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Pyruvoyl-dependent arginine decarboxylase | | Authors: | Tolbert, W.D, Graham, D.E, White, R.H, Ealick, S.E. | | Deposit date: | 2002-10-23 | | Release date: | 2003-03-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pyruvoyl-Dependent Arginine Decarboxylase from Methanococcus jannaschii:
Crystal Structures of the Self-Cleaved and S53A Proenzyme Forms
Structure, 11, 2003
|
|
1PR5
 
 | | Escherichia coli Purine Nucleoside Phosphorylase Complexed with 7-deazaadenosine and Phosphate/Sulfate | | Descriptor: | '2-(4-AMINO-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, PHOSPHATE ION, Purine nucleoside phosphorylase DeoD-type | | Authors: | Bennett, E.M, Li, C, Allan, P.W, Parker, W.B, Ealick, S.E. | | Deposit date: | 2003-06-19 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for substrate specificity of Escherichia coli purine nucleoside phosphorylase.
J.Biol.Chem., 278, 2003
|
|
1PR0
 
 | | Escherichia coli Purine Nucleoside Phosphorylase Complexed with Inosine and Phosphate/Sulfate | | Descriptor: | INOSINE, PHOSPHATE ION, Purine nucleoside phosphorylase DeoD-type | | Authors: | Bennett, E.M, Li, C, Allan, P.W, Parker, W.B, Ealick, S.E. | | Deposit date: | 2003-06-19 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for substrate specificity of Escherichia coli purine nucleoside phosphorylase.
J.Biol.Chem., 278, 2003
|
|
1MHM
 
 | |
1PBN
 
 | | PURINE NUCLEOSIDE PHOSPHORYLASE | | Descriptor: | PURINE NUCLEOSIDE PHOSPHORYLASE | | Authors: | Mao, C, Ealick, S.E. | | Deposit date: | 1995-07-10 | | Release date: | 1995-11-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Calf spleen purine nucleoside phosphorylase complexed with substrates and substrate analogues.
Biochemistry, 37, 1998
|
|
