6XJ3
 
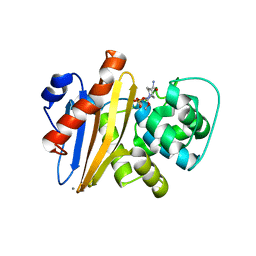 | | Crystal structure of Class D beta-lactamase from Klebsiella quasipneumoniae in complex with avibactam | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Chang, C, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-06-22 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Class D beta-lactamase from Klebsiella quasipneumoniae
To Be Published
|
|
4Q82
 
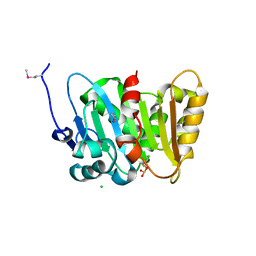 | | Crystal Structure of Phospholipase/Carboxylesterase from Haliangium ochraceum | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Kim, Y, Hatzos-Skintges, C, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-04-25 | | Release date: | 2014-05-14 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Crystal Structure of Phospholipase/Carboxylesterase from Haliangium ochraceum
To be Published
|
|
4Q2B
 
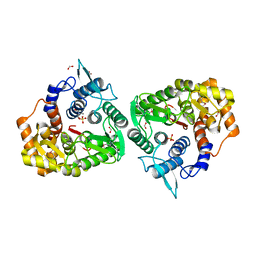 | | The crystal structure of an endo-1,4-D-glucanase from Pseudomonas putida KT2440 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endo-1,4-beta-D-glucanase, FORMIC ACID, ... | | Authors: | Tan, K, Joachimiak, G, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-04-07 | | Release date: | 2014-06-25 | | Last modified: | 2015-04-29 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | The crystal structure of an endo-1,4-D-glucanase from Pseudomonas putida KT2440
To be Published
|
|
4Q88
 
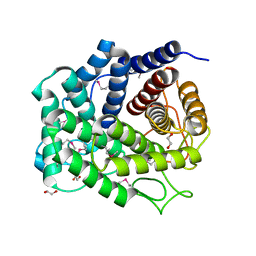 | | Glycosyl hydrolase family 88 from Bacteroides vulgatus | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Uncharacterized protein | | Authors: | Osipiuk, J, Li, H, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-04-25 | | Release date: | 2014-05-21 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Glycosyl hydrolase Family 88 from Bacteroides vulgatus
To be Published
|
|
4RGI
 
 | | Crystal Structure of KTSC Domain Protein YPO2434 from Yersinia pestis | | Descriptor: | GLYCEROL, SULFATE ION, Uncharacterized protein | | Authors: | Kim, Y, Chhor, G, Endres, M, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-09-30 | | Release date: | 2014-12-31 | | Method: | X-RAY DIFFRACTION (1.732 Å) | | Cite: | Crystal Structure of KTSC Domain Protein YPO2434 from Yersinia pestis
To be Published
|
|
6U7L
 
 | | 2.75 Angstrom Crystal Structure of Galactarate Dehydratase from Escherichia coli. | | Descriptor: | CALCIUM ION, CHLORIDE ION, Galactarate dehydratase (L-threo-forming) | | Authors: | Minasov, G, Shuvalova, L, Wawrzak, Z, Dubrovska, I, Kiryukhina, O, Endres, M, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-09-03 | | Release date: | 2019-11-06 | | Last modified: | 2021-01-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of galactarate dehydratase, a new fold in an enolase involved in bacterial fitness after antibiotic treatment.
Protein Sci., 29, 2020
|
|
6UAH
 
 | | Crystal Structure of the Metallo-beta-Lactamase L1 from Stenotrophomonas maltophilia in the Complex with Hydrolyzed Meropenem | | Descriptor: | (2~{S},3~{R},4~{S})-2-[(2~{S},3~{R})-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[(3~{S},5~{S})-5-(dimethylcarbamoy l)pyrrolidin-3-yl]sulfanyl-3-methyl-3,4-dihydro-2~{H}-pyrrole-5-carboxylic acid, 1,2-ETHANEDIOL, Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase), ... | | Authors: | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-09-10 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal Structure of the Metallo-beta-Lactamase L1 from Stenotrophomonas maltophilia in the Complex with Hydrolyzed Meropenem
To Be Published
|
|
4MDY
 
 | |
6UUK
 
 | | Crystal structure of muramoyltetrapeptide carboxypeptidase from Oxalobacter formigenes | | Descriptor: | Muramoyltetrapeptide carboxypeptidase | | Authors: | Chang, C, Tesar, C, Endres, M, Babnigg, G, Hassan, H, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2019-10-30 | | Release date: | 2020-11-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.348 Å) | | Cite: | Crystal structure of muramoyltetrapeptide carboxypeptidase from Oxalobacter formigenes
To Be Published
|
|
4LPQ
 
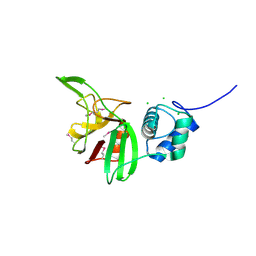 | | Crystal structure of the L,D-transpeptidase (residues 123-326) from Xylanimonas cellulosilytica DSM 15894 | | Descriptor: | CHLORIDE ION, ErfK/YbiS/YcfS/YnhG family protein | | Authors: | Nocek, B, Bigelow, L, Endres, M, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-07-16 | | Release date: | 2013-11-13 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Crystal structure of the L,D-transpeptidase (residues 123-326) from Xylanimonas cellulosilytica DSM 15894
To be Published
|
|
6V3Q
 
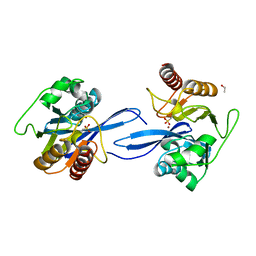 | | Crystal Structure of the Metallo-beta-Lactamase FIM-1 from Pseudomonas aeruginosa in the Mono-Zinc Form | | Descriptor: | ISOPROPYL ALCOHOL, Metallo-beta-lactamase FIM-1, ZINC ION | | Authors: | Kim, Y, Hatzos-Skintges, C, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-11-26 | | Release date: | 2020-01-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Metallo-beta-Lactamase FIM-1 from Pseudomonas aeruginosa in the Mono-Zinc Form
To Be Published
|
|
4MX8
 
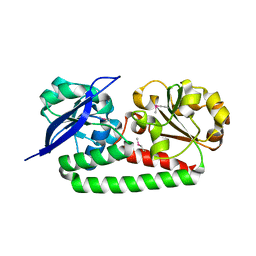 | |
4MVE
 
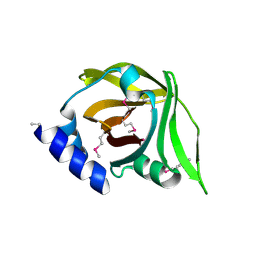 | |
6W6Y
 
 | | Crystal Structure of ADP ribose phosphatase of NSP3 from SARS CoV-2 in complex with AMP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADENOSINE MONOPHOSPHATE, Non-structural protein 3 | | Authors: | Michalska, K, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Mesecar, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-03-18 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | Crystal structures of SARS-CoV-2 ADP-ribose phosphatase: from the apo form to ligand complexes.
Iucrj, 7, 2020
|
|
6W02
 
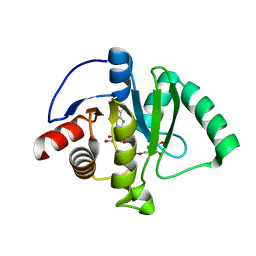 | | Crystal Structure of ADP ribose phosphatase of NSP3 from SARS CoV-2 in the complex with ADP ribose | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5-DIPHOSPHORIBOSE, Non-structural protein 3 | | Authors: | Michalska, K, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Mesecar, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-02-28 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of SARS-CoV-2 ADP-ribose phosphatase: from the apo form to ligand complexes.
Iucrj, 7, 2020
|
|
6VXS
 
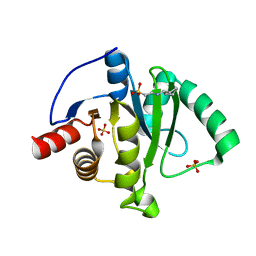 | | Crystal Structure of ADP ribose phosphatase of NSP3 from SARS CoV-2 | | Descriptor: | 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Non-structural protein 3, ... | | Authors: | Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Mesecar, A, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-02-24 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structures of SARS-CoV-2 ADP-ribose phosphatase: from the apo form to ligand complexes.
Iucrj, 7, 2020
|
|
6W9C
 
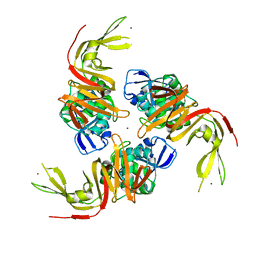 | | The crystal structure of papain-like protease of SARS CoV-2 | | Descriptor: | CHLORIDE ION, Non-structural protein 3, ZINC ION | | Authors: | Osipiuk, J, Jedrzejczak, R, Tesar, C, Endres, M, Stols, L, Babnigg, G, Kim, Y, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-03-22 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of papain-like protease of SARS CoV-2
to be published
|
|
6WCF
 
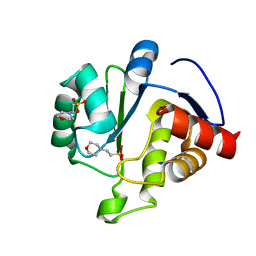 | | Crystal Structure of ADP ribose phosphatase of NSP3 from SARS-CoV-2 in complex with MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Non-structural protein 3 | | Authors: | Michalska, K, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Mesecar, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-03-30 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.065 Å) | | Cite: | Crystal structures of SARS-CoV-2 ADP-ribose phosphatase: from the apo form to ligand complexes.
Iucrj, 7, 2020
|
|
4OVX
 
 | | Crystal structure of Xylose isomerase domain protein from Planctomyces limnophilus DSM 3776 | | Descriptor: | 1,2-ETHANEDIOL, Xylose isomerase domain protein TIM barrel | | Authors: | Chang, C, Bigelow, L, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-01-22 | | Release date: | 2014-02-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.253 Å) | | Cite: | Crystal structure of Xylose isomerase domain protein from Planctomyces limnophilus DSM 3776
To be published
|
|
4PZJ
 
 | | 1.60 Angstrom resolution crystal structure of a transcriptional regulator of the LysR family from Eggerthella lenta DSM 2243 | | Descriptor: | CHLORIDE ION, Transcriptional regulator, LysR family | | Authors: | Halavaty, A.S, Filippova, E.V, Minasov, G, Kiryukhina, O, Endres, M, Shuvalova, L, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-03-31 | | Release date: | 2014-04-23 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 1.60 Angstrom resolution crystal structure of a transcriptional regulator of the LysR family from Eggerthella lenta DSM 2243
To be Published
|
|
4PYR
 
 | | Structure of a putative branched-chain amino acid ABC transporter from Chromobacterium violaceum ATCC 12472 | | Descriptor: | GLUTATHIONE, Putative branched-chain amino acid ABC transporter | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Endres, M, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-03-27 | | Release date: | 2014-04-23 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of a putative branched-chain amino acid ABC transporter from Chromobacterium violaceum ATCC 12472
To be Published
|
|
4Q6B
 
 | | Crystal Structure of ABC Transporter Substrate-Binding Protein fromDesulfitobacterium hafniense complex with Leu | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Extracellular ligand-binding receptor, ... | | Authors: | Kim, Y, Chhor, G, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-04-22 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.667 Å) | | Cite: | Crystal Structure of ABC Transporter Substrate-Binding Protein fromDesulfitobacterium hafniense complex with Leu
To be Published, 2014
|
|
4LQB
 
 | | Crystal structure of uncharacterized protein Kfla3161 | | Descriptor: | CITRIC ACID, GLYCEROL, Uncharacterized protein | | Authors: | Chang, C, Chhor, G, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-07-17 | | Release date: | 2013-07-31 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structure of uncharacterized protein Kfla3161
To be Published
|
|
4O5A
 
 | | The crystal structure of a LacI family transcriptional regulator from Bifidobacterium animalis subsp. lactis DSM 10140 | | Descriptor: | GLYCEROL, LacI family transcription regulator, SULFATE ION | | Authors: | Tan, K, Li, H, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-12-19 | | Release date: | 2014-01-15 | | Method: | X-RAY DIFFRACTION (1.777 Å) | | Cite: | The crystal structure of a LacI family transcriptional regulator from Bifidobacterium animalis subsp. lactis DSM 10140.
To be Published
|
|
4OVK
 
 | |
