6MHC
 
 | | Glutathione S-Transferase Omega 1 bound to covalent inhibitor 37 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2018-09-17 | | Release date: | 2019-02-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Design of N-(5-Phenylthiazol-2-yl)acrylamides as Novel and Potent Glutathione S-Transferase Omega 1 Inhibitors.
J. Med. Chem., 62, 2019
|
|
6MLQ
 
 | | Cryo-EM structure of microtubule-bound Kif7 in the ADP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Mani, N, Jiang, S, Wilson-Kubalek, E.M, Ku, P, Milligan, R.A, Subramanian, R. | | Deposit date: | 2018-09-27 | | Release date: | 2019-05-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Interplay between the Kinesin and Tubulin Mechanochemical Cycles Underlies Microtubule Tip Tracking by the Non-motile Ciliary Kinesin Kif7.
Dev.Cell, 49, 2019
|
|
6BVV
 
 | | Nipah virus W protein C-terminus in complex with Importin alpha 3 | | Descriptor: | Importin subunit alpha-3, Protein W | | Authors: | Smith, K.M, Tsimbalyuk, S, Edwards, M.R, Aragao, D, Cross, E.M, Basler, C.F, Forwood, J.K. | | Deposit date: | 2017-12-13 | | Release date: | 2018-07-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for importin alpha 3 specificity of W proteins in Hendra and Nipah viruses.
Nat Commun, 9, 2018
|
|
3JD5
 
 | | Cryo-EM structure of the small subunit of the mammalian mitochondrial ribosome | | Descriptor: | 28S ribosomal RNA, mitochondial, 28S ribosomal protein S10, ... | | Authors: | Kaushal, P.S, Sharma, M.R, Booth, T.M, Haque, E.M, Tung, C.S, Sanbonmatsu, K.Y, Spremulli, L.L, Agrawal, R.K. | | Deposit date: | 2016-04-08 | | Release date: | 2016-07-13 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Cryo-EM structure of the small subunit of the mammalian mitochondrial ribosome.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6N0K
 
 | | The complex of CCG-257081 bound to pirin | | Descriptor: | (3R)-N-(4-chlorophenyl)-5,5-difluoro-1-[3-fluoro-5-(pyridin-4-yl)benzene-1-carbonyl]piperidine-3-carboxamide, 1,2-ETHANEDIOL, FE (III) ION, ... | | Authors: | Lisabeth, E.M, Jin, X, Neubig, R. | | Deposit date: | 2018-11-07 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Identification of Pirin as a Molecular Target of the CCG-1423/CCG-203971 Series of Antifibrotic and Antimetastatic Compounds
ACS Pharmacol Transl Sci, 2, 2019
|
|
6C1G
 
 | | High-Resolution Cryo-EM Structures of Actin-bound Myosin States Reveal the Mechanism of Myosin Force Sensing | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Mentes, A, Huehn, A, Liu, X, Zwolak, A, Dominguez, R, Shuman, H, Ostap, E.M, Sindelar, C.V. | | Deposit date: | 2018-01-04 | | Release date: | 2018-01-31 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | High-resolution cryo-EM structures of actin-bound myosin states reveal the mechanism of myosin force sensing.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6BVT
 
 | | Importin alpha 1 in cargo free state | | Descriptor: | Importin subunit alpha-1 | | Authors: | Smith, K.M, Tsimablyuk, S, Edwards, M.R, Aragao, D, Cross, E.M, Basler, C.F, Forwood, J.K. | | Deposit date: | 2017-12-13 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for importin alpha 3 specificity of W proteins in Hendra and Nipah viruses.
Nat Commun, 9, 2018
|
|
6CJG
 
 | | Human dihydroorotate dehydrogenase bound to napthyridine inhibitor 46 | | Descriptor: | 2-([1,1'-biphenyl]-4-yl)-3-methyl-1,7-naphthyridine-4-carboxylic acid, 3-[decyl(dimethyl)ammonio]propane-1-sulfonate, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2018-02-26 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.851 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of 4-Quinoline Carboxylic Acids as Inhibitors of Dihydroorotate Dehydrogenase.
J. Med. Chem., 61, 2018
|
|
6BW9
 
 | | Hendra virus W protein C-terminus in complex with Importin alpha 3 crystal form 1 | | Descriptor: | Importin subunit alpha-3, Protein W | | Authors: | Tsimbalyuk, S, Smith, K.M, Edwards, M.R, Aragao, D, Cross, E.M, Basler, C.F, Forwood, J.K. | | Deposit date: | 2017-12-14 | | Release date: | 2018-07-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for importin alpha 3 specificity of W proteins in Hendra and Nipah viruses.
Nat Commun, 9, 2018
|
|
6C1D
 
 | | High-Resolution Cryo-EM Structures of Actin-bound Myosin States Reveal the Mechanism of Myosin Force Sensing | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Mentes, A, Huehn, A, Liu, X, Zwolak, A, Dominguez, R, Shuman, H, Ostap, E.M, Sindelar, C.V. | | Deposit date: | 2018-01-04 | | Release date: | 2018-01-31 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | High-resolution cryo-EM structures of actin-bound myosin states reveal the mechanism of myosin force sensing.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6BWA
 
 | | Hendra virus W protein C-terminus in complex with Importin alpha 3 crystal form 2 | | Descriptor: | Importin subunit alpha-3, Protein W | | Authors: | Tsimbalyuk, S, Smith, K.M, Edwards, M.R, Aragao, D, Cross, E.M, Basler, C.F, Forwood, J.K. | | Deposit date: | 2017-12-14 | | Release date: | 2018-07-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for importin alpha 3 specificity of W proteins in Hendra and Nipah viruses.
Nat Commun, 9, 2018
|
|
6BVZ
 
 | | Importin alpha 3 in cargo free state | | Descriptor: | Importin subunit alpha-3 | | Authors: | Smith, K.M, Tsimbalyuk, S, Edwards, M.R, Aragao, D, Cross, E.M, Basler, C.F, Forwood, J.K. | | Deposit date: | 2017-12-14 | | Release date: | 2018-07-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for importin alpha 3 specificity of W proteins in Hendra and Nipah viruses.
Nat Commun, 9, 2018
|
|
6C1H
 
 | | High-Resolution Cryo-EM Structures of Actin-bound Myosin States Reveal the Mechanism of Myosin Force Sensing | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Mentes, A, Huehn, A, Liu, X, Zwolak, A, Dominguez, R, Shuman, H, Ostap, E.M, Sindelar, C.V. | | Deposit date: | 2018-01-04 | | Release date: | 2018-01-31 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | High-resolution cryo-EM structures of actin-bound myosin states reveal the mechanism of myosin force sensing.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3KVO
 
 | | Crystal structure of the catalytic domain of human Hydroxysteroid dehydrogenase like 2 (HSDL2) | | Descriptor: | Hydroxysteroid dehydrogenase-like protein 2, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ugochukwu, E, Bhatia, C, Huang, J, Pilka, E, Muniz, J.R.C, Pike, A.C.W, Krojer, T, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Bountra, C, Verdin, E.M, Oppermann, U, Kavanagh, K.L, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-11-30 | | Release date: | 2010-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the catalytic domain of human Hydroxysteroid dehydrogenase like 2 (HSDL2)
To be Published
|
|
6NRP
 
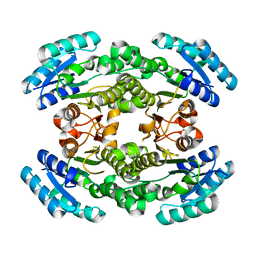 | | Putative short-chain dehydrogenase/reductase (SDR) from Acinetobacter baumannii | | Descriptor: | 3-oxoacyl-ACP reductase FabG | | Authors: | Cross, E.M, Smith, K.M, Shaw, K.I, Aragao, D, Forwood, J.K. | | Deposit date: | 2019-01-23 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights into Acinetobacter baumannii fatty acid synthesis 3-oxoacyl-ACP reductases.
Sci Rep, 11, 2021
|
|
3IVK
 
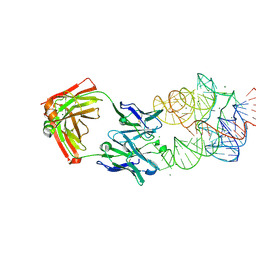 | | Crystal Structure of the Catalytic Core of an RNA Polymerase Ribozyme Complexed with an Antigen Binding Antibody Fragment | | Descriptor: | CADMIUM ION, CHLORIDE ION, Fab heavy chain, ... | | Authors: | Koldobskaya, Y, Duguid, E.M, Shechner, D.M, Koide, S, Kossiakoff, A.A, Bartel, D.P, Piccirilli, J.A. | | Deposit date: | 2009-09-01 | | Release date: | 2010-03-02 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of the catalytic core of an RNA-polymerase ribozyme.
Science, 326, 2009
|
|
3IVT
 
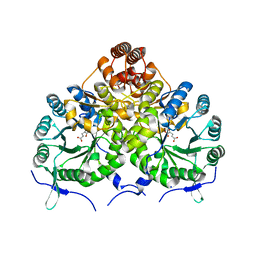 | | Homocitrate Synthase Lys4 bound to 2-OG | | Descriptor: | 2-OXOGLUTARIC ACID, Homocitrate synthase, mitochondrial, ... | | Authors: | Bulfer, S.L, Scott, E.M, Couture, J.-F, Pillus, L, Trievel, R.C. | | Deposit date: | 2009-09-01 | | Release date: | 2009-09-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Crystal structure and functional analysis of homocitrate synthase, an essential enzyme in lysine biosynthesis.
J.Biol.Chem., 284, 2009
|
|
6CJF
 
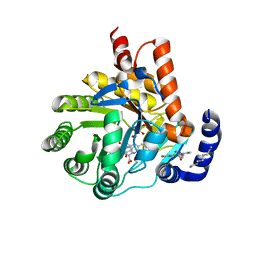 | | Human dihydroorotate dehydrogenase bound to 4-quinoline carboxylic acid inhibitor 43 | | Descriptor: | 2-[4-(2-chloro-6-methylpyridin-3-yl)phenyl]-6-fluoro-3-methylquinoline-4-carboxylic acid, 3-[decyl(dimethyl)ammonio]propane-1-sulfonate, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2018-02-26 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of 4-Quinoline Carboxylic Acids as Inhibitors of Dihydroorotate Dehydrogenase.
J. Med. Chem., 61, 2018
|
|
6BW1
 
 | | Hendra virus W protein C-terminus in complex with Importin alpha 1 | | Descriptor: | Importin subunit alpha-1, Protein W | | Authors: | Tsimbalyuk, S, Smith, K.M, Edwards, M.R, Aragao, D, Cross, E.M, Basler, C.F, Forwood, J.K. | | Deposit date: | 2017-12-14 | | Release date: | 2018-07-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for importin alpha 3 specificity of W proteins in Hendra and Nipah viruses.
Nat Commun, 9, 2018
|
|
6BW0
 
 | | Nipah virus W protein C-terminus in complex with Importin alpha 1 | | Descriptor: | Importin subunit alpha-1, Protein W | | Authors: | Smith, K.M, Tsimbalyuk, S, Edwards, M.R, Aragao, D, Cross, E.M, Basler, C.F, Forwood, J.K. | | Deposit date: | 2017-12-14 | | Release date: | 2018-07-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for importin alpha 3 specificity of W proteins in Hendra and Nipah viruses.
Nat Commun, 9, 2018
|
|
6BWB
 
 | | Hendra virus W protein C-terminus in complex with Importin alpha 3 crystal form 3 | | Descriptor: | Importin subunit alpha-3, Protein W | | Authors: | Tsimbalyuk, S, Smith, K.M, Edwards, M.R, Aragao, D, Cross, E.M, Basler, C.F, Forwood, J.K. | | Deposit date: | 2017-12-14 | | Release date: | 2018-07-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for importin alpha 3 specificity of W proteins in Hendra and Nipah viruses.
Nat Commun, 9, 2018
|
|
6ONP
 
 | |
6ASZ
 
 | |
6DS2
 
 | | Crystal structure of Ni(II)-bound human calprotectin | | Descriptor: | NICKEL (II) ION, Protein S100-A8, Protein S100-A9, ... | | Authors: | Nolan, E.M, Drennan, C.L, Nakashige, T.G. | | Deposit date: | 2018-06-13 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biophysical Examination of the Calcium-Modulated Nickel-Binding Properties of Human Calprotectin Reveals Conformational Change in the EF-Hand Domains and His3Asp Site.
Biochemistry, 57, 2018
|
|
3L08
 
 | | Structure of Pi3K gamma with a potent inhibitor: GSK2126458 | | Descriptor: | 2,4-difluoro-N-[2-methoxy-5-(4-pyridazin-4-ylquinolin-6-yl)pyridin-3-yl]benzenesulfonamide, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Elkins, P.A, Marrero, E.M. | | Deposit date: | 2009-12-09 | | Release date: | 2010-06-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of GSK2126458, a Highly Potent Inhibitor of PI3K and the Mammalian Target of Rapamycin.
ACS Med Chem Lett, 1, 2010
|
|
