1PK9
 
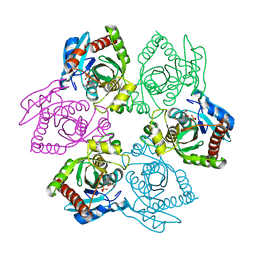 | | Crystal Structure of E. coli purine nucleoside phosphorylase complexed with 2-fluoroadenosine and sulfate/phosphate | | 分子名称: | 2-(6-AMINO-2-FLUORO-PURIN-9-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, PHOSPHATE ION, Purine nucleoside phosphorylase DeoD-type | | 著者 | Bennett, E.M, Li, C, Allan, P.W, Parker, W.B, Ealick, S.E. | | 登録日 | 2003-06-05 | | 公開日 | 2003-11-25 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural basis for substrate specificity of Escherichia coli purine nucleoside phosphorylase.
J.Biol.Chem., 278, 2003
|
|
1PKE
 
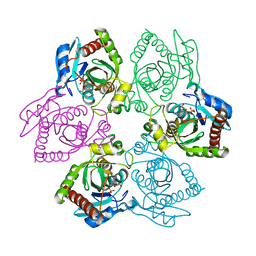 | | Crystal Structure of E. coli purine nucleoside phosphorylase complexed with 2-fluoro-2'-deoxyadenosine and sulfate/phosphate | | 分子名称: | 5-(6-AMINO-2-FLUORO-PURIN-9-YL)-2-HYDROXYMETHYL-TETRAHYDRO-FURAN-3-OL, PHOSPHATE ION, Purine nucleoside phosphorylase DeoD-type | | 著者 | Bennett, E.M, Li, C, Allan, P.W, Parker, W.B, Ealick, S.E. | | 登録日 | 2003-06-05 | | 公開日 | 2003-11-25 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural basis for substrate specificity of Escherichia coli purine nucleoside phosphorylase.
J.Biol.Chem., 278, 2003
|
|
1TRE
 
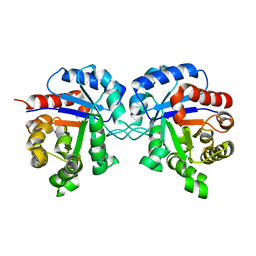 | |
1U56
 
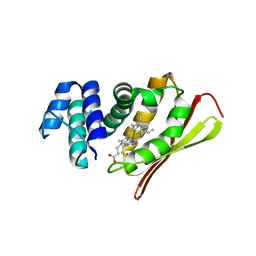 | | Crystal structure of an oxygen binding H-NOX domain related to soluble guanylate cyclases (Water-ligated, ferric form) | | 分子名称: | CHLORIDE ION, Heme-based Methyl-accepting chemotaxis protein, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Pellicena, P, Karow, D.S, Boon, E.M, Marletta, M.A, Kuriyan, J. | | 登録日 | 2004-07-27 | | 公開日 | 2004-08-31 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of an oxygen-binding heme domain related to soluble guanylate cyclases.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1TPE
 
 | |
1PW7
 
 | | Crystal Structure of E. coli purine nucleoside phosphorylase complexed with 9-beta-D-arabinofuranosyladenine and sulfate/phosphate | | 分子名称: | 2-(6-AMINO-PURIN-9-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, PHOSPHATE ION, Purine nucleoside phosphorylase DeoD-type | | 著者 | Bennett, E.M, Li, C, Allan, P.W, Parker, W.B, Ealick, S.E. | | 登録日 | 2003-06-30 | | 公開日 | 2003-11-25 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural basis for substrate specificity of Escherichia coli purine nucleoside phosphorylase.
J.Biol.Chem., 278, 2003
|
|
1PR4
 
 | | Escherichia coli Purine Nucleoside Phosphorylase Complexed with 9-beta-D-ribofuranosyl-6-methylthiopurine and Phosphate/Sulfate | | 分子名称: | 2-HYDROXYMETHYL-5-(6-METHYLSULFANYL-PURIN-9-YL)-TETRAHYDRO-FURAN-3,4-DIOL, PHOSPHATE ION, Purine nucleoside phosphorylase DeoD-type | | 著者 | Bennett, E.M, Li, C, Allan, P.W, Parker, W.B, Ealick, S.E. | | 登録日 | 2003-06-19 | | 公開日 | 2003-11-25 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural basis for substrate specificity of Escherichia coli purine nucleoside phosphorylase.
J.Biol.Chem., 278, 2003
|
|
2N9P
 
 | | Solution structure of RNF126 N-terminal zinc finger domain in complex with BAG6 Ubiquitin-like domain | | 分子名称: | E3 ubiquitin-protein ligase RNF126, Large proline-rich protein BAG6, ZINC ION | | 著者 | Martinez-Lumbreras, S, Krysztofinska, E.M, Thapaliya, A, Isaacson, R.L. | | 登録日 | 2015-12-01 | | 公開日 | 2016-05-25 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural and functional insights into the E3 ligase, RNF126.
Sci Rep, 6, 2016
|
|
2N34
 
 | | NMR assignments and solution structure of the JAK interaction region of SOCS5 | | 分子名称: | Suppressor of cytokine signaling 5 | | 著者 | Chandrashekaran, I.R, Mohanty, B, Linossi, E.M, Nicholson, S.E, Babon, J, Norton, R.S, Dagley, L.F, Leung, E.W.W, Murphy, J.M. | | 登録日 | 2015-05-21 | | 公開日 | 2015-07-29 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure and Functional Characterization of the Conserved JAK Interaction Region in the Intrinsically Disordered N-Terminus of SOCS5.
Biochemistry, 54, 2015
|
|
1U0X
 
 | | Crystal structure of nitrophorin 4 under pressure of xenon (200 psi) | | 分子名称: | AMMONIA, Nitrophorin 4, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Nienhaus, K, Maes, E.M, Weichsel, A, Montfort, W.R, Nienhaus, G.U. | | 登録日 | 2004-07-14 | | 公開日 | 2004-07-20 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Structural dynamics controls nitric oxide affinity in nitrophorin 4
J.Biol.Chem., 279, 2004
|
|
1U4H
 
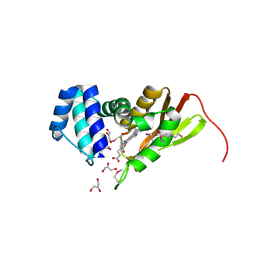 | | Crystal structure of an oxygen binding H-NOX domain related to soluble guanylate cyclases (oxygen complex) | | 分子名称: | GLYCEROL, Heme-based Methyl-accepting Chemotaxis Protein, OXYGEN MOLECULE, ... | | 著者 | Pellicena, P, Karow, D.S, Boon, E.M, Marletta, M.A, Kuriyan, J. | | 登録日 | 2004-07-25 | | 公開日 | 2004-08-31 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | Crystal structure of an oxygen-binding heme domain related to soluble guanylate cyclases.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1U8T
 
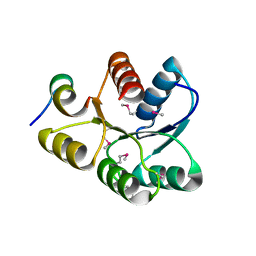 | | Crystal structure of CheY D13K Y106W alone and in complex with a FliM peptide | | 分子名称: | Chemotaxis protein cheY, Flagellar motor switch protein fliM, SULFATE ION | | 著者 | Dyer, C.M, Quillin, M.L, Campos, A, Lu, J, McEvoy, M.M, Hausrath, A.C, Westbrook, E.M, Matsumura, P, Matthews, B.W, Dahlquist, F.W. | | 登録日 | 2004-08-06 | | 公開日 | 2004-10-05 | | 最終更新日 | 2021-10-20 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structure of the Constitutively Active Double Mutant CheY(D13K Y106W) Alone and in Complex with a FliM Peptide
J.Mol.Biol., 342, 2004
|
|
2MZZ
 
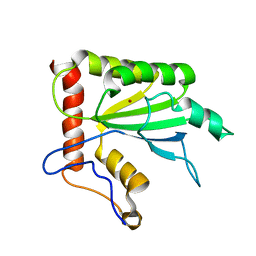 | | NMR structure of APOBEC3G NTD variant, sNTD | | 分子名称: | Apolipoprotein B mRNA-editing enzyme, catalytic polypeptide-like 3G variant, ZINC ION | | 著者 | Kouno, T, Luengas, E.M, Shigematu, M, Shandilya, S.M.D, Zhang, J, Chen, L, Hara, M, Schiffer, C.A, Harris, R.S, Matsuo, H. | | 登録日 | 2015-02-28 | | 公開日 | 2015-05-13 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of the Vif-binding domain of the antiviral enzyme APOBEC3G.
Nat.Struct.Mol.Biol., 22, 2015
|
|
1NZ6
 
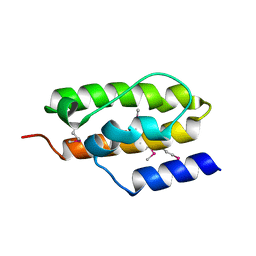 | | Crystal Structure of Auxilin J-Domain | | 分子名称: | Auxilin | | 著者 | Jiang, J, Taylor, A.B, Prasad, K, Ishikawa-Brush, Y, Hart, P.J, Lafer, E.M, Sousa, R. | | 登録日 | 2003-02-16 | | 公開日 | 2003-04-22 | | 最終更新日 | 2011-11-16 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure-function analysis of the auxilin J-domain reveals an extended Hsc70 interaction interface.
Biochemistry, 42, 2003
|
|
1U55
 
 | | Crystal structure of an oxygen binding H-NOX domain related to soluble guanylate cyclases (oxygen complex) | | 分子名称: | CHLORIDE ION, Heme-based Methyl-accepting Chemotaxis Protein, OXYGEN MOLECULE, ... | | 著者 | Pellicena, P, Karow, D.S, Boon, E.M, Marletta, M.A, Kuriyan, J. | | 登録日 | 2004-07-27 | | 公開日 | 2004-08-31 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Crystal structure of an oxygen-binding heme domain related to soluble guanylate cyclases.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
2N5Y
 
 | | Solution NMR structure of octyl-tridecaptin A1 in DPC micelles containing Gram-negative lipid II | | 分子名称: | Octyl-tridecaptin A1 | | 著者 | Cochrane, S.A, Findlay, B, Bakhtiary, A, Rodriguez-Lopez, E.M, Vederas, J.C. | | 登録日 | 2015-08-03 | | 公開日 | 2016-09-28 | | 最終更新日 | 2023-11-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Antimicrobial lipopeptide tridecaptin A1 selectively binds to Gram-negative lipid II.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1TZ8
 
 | | The monoclinic crystal structure of transthyretin in complex with diethylstilbestrol | | 分子名称: | ACETATE ION, CARBONATE ION, DIETHYLSTILBESTROL, ... | | 著者 | Morais-de-Sa, E.M, Pereira, P.J.B, Saraiva, M.J, Damas, A.M. | | 登録日 | 2004-07-09 | | 公開日 | 2004-10-12 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | The crystal structure of transthyretin in complex with diethylstilbestrol: a promising template for the design of amyloid inhibitors
J.Biol.Chem., 279, 2004
|
|
1U0F
 
 | | Crystal structure of mouse phosphoglucose isomerase in complex with glucose 6-phosphate | | 分子名称: | 6-O-phosphono-alpha-D-glucopyranose, BETA-MERCAPTOETHANOL, GLUCOSE-6-PHOSPHATE, ... | | 著者 | Solomons, J.T.G, Zimmerly, E.M, Burns, S, Krishnamurthy, N, Swan, M.K, Krings, S, Muirhead, H, Chirgwin, J, Davies, C. | | 登録日 | 2004-07-13 | | 公開日 | 2004-11-02 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | The crystal structure of mouse phosphoglucose isomerase at 1.6A resolution and its complex with glucose 6-phosphate reveals the catalytic mechanism of sugar ring opening.
J.Mol.Biol., 342, 2004
|
|
1U0G
 
 | | Crystal structure of mouse phosphoglucose isomerase in complex with erythrose 4-phosphate | | 分子名称: | BETA-MERCAPTOETHANOL, ERYTHOSE-4-PHOSPHATE, GLYCEROL, ... | | 著者 | Solomons, J.T.G, Zimmerly, E.M, Burns, S, Krishnamurthy, N, Swan, M.K, Krings, S, Muirhead, H, Chirgwin, J, Davies, C. | | 登録日 | 2004-07-13 | | 公開日 | 2004-11-02 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | The crystal structure of mouse phosphoglucose isomerase at 1.6A resolution and its complex with glucose 6-phosphate reveals the catalytic mechanism of sugar ring opening.
J.Mol.Biol., 342, 2004
|
|
1OIY
 
 | | Structure of human Thr160-phospho CDK2/cyclin A complexed with a 6-cyclohexylmethyloxy-2-anilino-purine inhibitor | | 分子名称: | 4-(6-CYCLOHEXYLMETHOXY-9H-PURIN-2-YLAMINO)--BENZAMIDE, CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, ... | | 著者 | Pratt, D.J, Endicott, J.A, Noble, M.E.M. | | 登録日 | 2003-06-26 | | 公開日 | 2004-07-13 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | N2-Substituted O6-Cyclohexylmethylguanine Derivatives: Potent Inhibitors of Cyclin-Dependent Kinases 1 and 2
J.Med.Chem., 47, 2004
|
|
1OI9
 
 | | Structure of human Thr160-phospho CDK2/cyclin A complexed with a 6-cyclohexylmethyloxy-2-anilino-purine inhibitor | | 分子名称: | 6-CYCLOHEXYLMETHYLOXY-2-(4'-HYDROXYANILINO)PURINE, CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, ... | | 著者 | Pratt, D.J, Endicott, J.A, Noble, M.E.M. | | 登録日 | 2003-06-10 | | 公開日 | 2004-07-13 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | N2-Substituted O6-Cyclohexylmethylguanine Derivatives: Potent Inhibitors of Cyclin-Dependent Kinases 1 and 2
J.Med.Chem., 47, 2004
|
|
1OIU
 
 | | Structure of human Thr160-phospho CDK2/cyclin A complexed with a 6-cyclohexylmethyloxy-2-anilino-purine inhibitor | | 分子名称: | 3-(6-CYCLOHEXYLMETHOXY-9H-PURIN-2-YLAMINO)-BENZENESULFONAMIDE, CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, ... | | 著者 | Pratt, D.J, Endicott, J.A, Noble, M.E.M. | | 登録日 | 2003-06-26 | | 公開日 | 2004-07-13 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | N2-Substituted O6-Cyclohexylmethylguanine Derivatives: Potent Inhibitors of Cyclin-Dependent Kinases 1 and 2
J.Med.Chem., 47, 2004
|
|
1TT6
 
 | | The orthorhombic crystal structure of transthyretin in complex with diethylstilbestrol | | 分子名称: | DIETHYLSTILBESTROL, GLYCEROL, SULFATE ION, ... | | 著者 | Morais-de-Sa, E.M, Pereira, P.J.B, Saraiva, M.J, Damas, A.M. | | 登録日 | 2004-06-22 | | 公開日 | 2004-10-12 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The crystal structure of transthyretin in complex with diethylstilbestrol: a promising template for the design of amyloid inhibitors
J.Biol.Chem., 279, 2004
|
|
1U0E
 
 | | Crystal structure of mouse phosphoglucose isomerase | | 分子名称: | BETA-MERCAPTOETHANOL, GLYCEROL, Glucose-6-phosphate isomerase, ... | | 著者 | Solomons, J.T.G, Zimmerly, E.M, Burns, S, Krishnamurthy, N, Swan, M.K, Krings, S, Muirhead, H, Chirgwin, J, Davies, C. | | 登録日 | 2004-07-13 | | 公開日 | 2004-11-02 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | The crystal structure of mouse phosphoglucose isomerase at 1.6A resolution and its complex with glucose 6-phosphate reveals the catalytic mechanism of sugar ring opening.
J.Mol.Biol., 342, 2004
|
|
2N9O
 
 | | Solution structure of RNF126 N-terminal zinc finger domain | | 分子名称: | E3 ubiquitin-protein ligase RNF126, ZINC ION | | 著者 | Martinez-Lumbreras, S, Krysztofinska, E.M, Thapaliya, A, Isaacson, R.L. | | 登録日 | 2015-12-01 | | 公開日 | 2016-05-25 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural and functional insights into the E3 ligase, RNF126.
Sci Rep, 6, 2016
|
|
