6FSZ
 
 | | Structure of the nuclear RNA exosome | | Descriptor: | ATP-dependent RNA helicase DOB1, Exosome complex component CSL4, Exosome complex component MTR3, ... | | Authors: | Schuller, J.M, Falk, S, Conti, E. | | Deposit date: | 2018-02-20 | | Release date: | 2018-03-21 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structure of the nuclear exosome captured on a maturing preribosome.
Science, 360, 2018
|
|
4U21
 
 | | GluA2flip sLBD complexed with FW and (R,R)-2b crystal form E | | Descriptor: | 2-AMINO-3-(5-FLUORO-2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, Glutamate receptor 2,Glutamate receptor 2, N,N'-[biphenyl-4,4'-diyldi(2R)propane-2,1-diyl]dipropane-2-sulfonamide | | Authors: | Chen, L, Gouaux, E. | | Deposit date: | 2014-07-16 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.3908 Å) | | Cite: | Structure and Dynamics of AMPA Receptor GluA2 in Resting, Pre-Open, and Desensitized States.
Cell, 158, 2014
|
|
6FUG
 
 | | Complement factor D in complex with the inhibitor 3-((3-((3-(aminomethyl)phenyl)amino)-1H-pyrazolo[3,4-d]pyrimidin-4-yl)amino)phenol | | Descriptor: | 3-[[3-[[3-(aminomethyl)phenyl]amino]-1~{H}-pyrazolo[3,4-d]pyrimidin-4-yl]amino]phenol, Complement factor D | | Authors: | Mac Sweeney, A, Ostermann, N, Vulpetti, A, Maibaum, J, Erbel, P, Lorthiois, E, Yoon, T, Randl, S, Ruedisser, S. | | Deposit date: | 2018-02-27 | | Release date: | 2018-06-06 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Discovery and Design of First Benzylamine-Based Ligands Binding to an Unlocked Conformation of the Complement Factor D.
ACS Med Chem Lett, 9, 2018
|
|
5F7L
 
 | | Blood group antigen binding adhesin BabA of Helicobacter pylori strain 17875 in complex with Nanobody Nb-ER14 | | Descriptor: | Adhesin binding fucosylated histo-blood group antigen, Nanobody Nb-ER14, SULFATE ION | | Authors: | Moonens, K, Gideonsson, P, Subedi, S, Romao, E, Oscarson, S, Muyldermans, S, Boren, T, Remaut, H. | | Deposit date: | 2015-12-08 | | Release date: | 2016-01-20 | | Last modified: | 2016-01-27 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structural Insights into Polymorphic ABO Glycan Binding by Helicobacter pylori.
Cell Host Microbe, 19, 2016
|
|
8DFO
 
 | | type I-C Cascade bound to AcrIC4 | | Descriptor: | AcrIC4, CRISPR-associated protein, CT1133 family, ... | | Authors: | O'Brien, R.E, Bravo, J.P.K, Ramos, D, Hibshman, G.N, Wright, J.T, Taylor, D.W. | | Deposit date: | 2022-06-22 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural snapshots of R-loop formation by a type I-C CRISPR Cascade.
Mol.Cell, 83, 2023
|
|
1G1O
 
 | | CRYSTAL STRUCTURE OF THE HIGHLY AMYLOIDOGENIC TRANSTHYRETIN MUTANT TTR G53S/E54D/L55S | | Descriptor: | TRANSTHYRETIN | | Authors: | Eneqvist, T, Andersson, K, Olofsson, A, Lundgren, E, Sauer-Eriksson, A.E. | | Deposit date: | 2000-10-13 | | Release date: | 2001-10-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The beta-slip: a novel concept in transthyretin amyloidosis.
Mol.Cell, 6, 2000
|
|
8DFA
 
 | | type I-C Cascade bound to ssDNA target | | Descriptor: | CRISPR-associated protein, CT1133 family, TM1801 family, ... | | Authors: | O'Brien, R.E, Bravo, J.P.K, Ramos, D, Hibshman, G.N, Wright, J.T, Taylor, D.W. | | Deposit date: | 2022-06-21 | | Release date: | 2023-02-22 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural snapshots of R-loop formation by a type I-C CRISPR Cascade.
Mol.Cell, 83, 2023
|
|
4ZKD
 
 | |
4ZHG
 
 | | Siderocalin-mediated recognition and cellular uptake of actinides | | Descriptor: | AMERICIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Allred, B.E, Rupert, P.B, Gauny, S.S, An, D.D, Ralston, C.Y, Sturzbecher-Hoehne, M, Strong, R.K, Abergel, R.J. | | Deposit date: | 2015-04-24 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Siderocalin-mediated recognition, sensitization, and cellular uptake of actinides.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4TV8
 
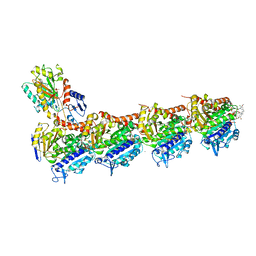 | | Tubulin-Maytansine complex | | Descriptor: | (3beta,4beta,5beta,10beta,11E,13E)-maytansine, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Prota, A.E, Bargsten, K, Diaz, J.F, Marsh, M, Cuevas, C, Liniger, M, Neuhaus, C, Andreu, J.M, Altmann, K.H, Steinmetz, M.O. | | Deposit date: | 2014-06-26 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | A new tubulin-binding site and pharmacophore for microtubule-destabilizing anticancer drugs.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2XX9
 
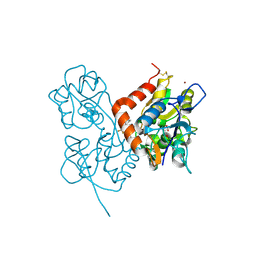 | | Crystal structure of 1-((2-fluoro-4-(3-(trifluoromethyl)-4,5,6,7- tetrahydro-1H-indazol-1-yl)phenyl)methyl)-2-pyrrolidinone in complex with the ligand binding domain of the Rat GluA2 receptor and glutamate at 2.2A resolution. | | Descriptor: | GLUTAMATE RECEPTOR 2, GLUTAMIC ACID, N,N-DIMETHYL-4-[3-(TRIFLUOROMETHYL)-4,5,6,7-TETRAHYDRO-1H-INDAZOL-1-YL]BENZAMIDE, ... | | Authors: | Ward, S.E, Harries, M, Aldegheri, L, Austin, N.E, Ballantine, S, Ballini, E, Bradley, D.M, Bax, B.D, Clarke, B.P, Harris, A.J, Harrison, S.A, Melarange, R.A, Mookherjee, C, Mosley, J, DalNegro, G, Oliosi, B, Smith, K.J, Thewlis, K.M, Woollard, P.M, Yusaf, S.P. | | Deposit date: | 2010-11-09 | | Release date: | 2011-04-06 | | Last modified: | 2011-09-28 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Integration of Lead Optimization with Crystallography for a Membrane-Bound Ion Channel Target: Discovery of a New Class of Ampa Receptor Positive Allosteric Modulators.
J.Med.Chem., 54, 2011
|
|
8DAJ
 
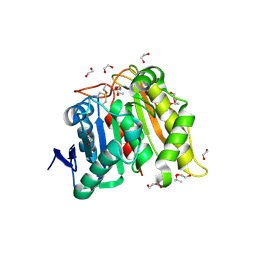 | | Structure and Biochemistry of a Promiscuous Thermophilic Polyhydroxybutyrate Depolymerase from Lihuaxuella thermophilia | | Descriptor: | 1,2-ETHANEDIOL, Esterase, PHB depolymerase family, ... | | Authors: | Thomas, G.M, Quirk, S, Huard, D.J.E, Lieberman, R.L. | | Deposit date: | 2022-06-13 | | Release date: | 2023-02-15 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Bioplastic degradation by a polyhydroxybutyrate depolymerase from a thermophilic soil bacterium.
Protein Sci., 31, 2022
|
|
8DEJ
 
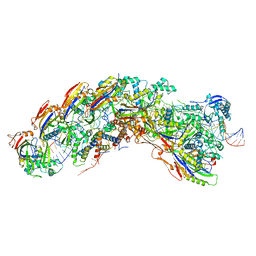 | | D. vulgaris type I-C Cascade bound to dsDNA target | | Descriptor: | CRISPR-associated protein, CT1133 family, TM1801 family, ... | | Authors: | O'Brien, R.E, Bravo, J.P.K, Ramos, D, Hibshman, G.N, Wright, J.T, Taylor, D.W. | | Deposit date: | 2022-06-20 | | Release date: | 2023-02-22 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structural snapshots of R-loop formation by a type I-C CRISPR Cascade.
Mol.Cell, 83, 2023
|
|
1RLO
 
 | | Phospho-aspartyl Intermediate Analogue of ybiV from E. coli K12 | | Descriptor: | GLYCEROL, MAGNESIUM ION, Phosphatase | | Authors: | Roberts, A, Lee, S.Y, McCullagh, E, Silversmith, R.E, Wemmer, D.E. | | Deposit date: | 2003-11-26 | | Release date: | 2004-12-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ybiv from Escherichia coli K12 is a HAD phosphatase.
Proteins, 58, 2005
|
|
8DEX
 
 | | type I-C Cascade | | Descriptor: | CRISPR-associated protein, CT1133 family, TM1801 family, ... | | Authors: | O'Brien, R.E, Bravo, J.P.K, Ramos, D, Hibshman, G.N, Wright, J.T, Taylor, D.W. | | Deposit date: | 2022-06-21 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural snapshots of R-loop formation by a type I-C CRISPR Cascade.
Mol.Cell, 83, 2023
|
|
2XX7
 
 | | Crystal structure of 1-(4-(1-pyrrolidinylcarbonyl)phenyl)-3-(trifluoromethyl)-4,5,6,7-tetrahydro-1H-indazole in complex with the ligand binding domain of the Rat GluA2 receptor and glutamate at 2.2A resolution. | | Descriptor: | 1-[4-(1-PYRROLIDINYLCARBONYL)PHENYL]-3-(TRIFLUOROMETHYL)-4,5,6,7-TETRAHYDRO-1H-INDAZOLE, GLUTAMATE RECEPTOR 2, GLUTAMIC ACID, ... | | Authors: | Ward, S.E, Harries, M, Aldegheri, L, Austin, N.E, Ballantine, S, Ballini, E, Bradley, D.M, Bax, B.D, Clarke, B.P, Harris, A.J, Harrison, S.A, Melarange, R.A, Mookherjee, C, Mosley, J, DalNegro, G, Oliosi, B, Smith, K.J, Thewlis, K.M, Woollard, P.M, Yusaf, S.P. | | Deposit date: | 2010-11-09 | | Release date: | 2011-04-06 | | Last modified: | 2011-09-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Integration of Lead Optimization with Crystallography for a Membrane-Bound Ion Channel Target: Discovery of a New Class of Ampa Receptor Positive Allosteric Modulators.
J.Med.Chem., 54, 2011
|
|
5J6T
 
 | |
5J6V
 
 | |
5J6W
 
 | |
2XXH
 
 | | Crystal structure of 1-(4-(2-oxo-2-(1-pyrrolidinyl)ethyl)phenyl)-3-(trifluoromethyl)-4,5,6,7-tetrahydro-1H-indazole in complex with the ligand binding domain of the Rat GluA2 receptor and glutamate at 1.5A resolution. | | Descriptor: | 1-{4-[2-OXO-2-(1-PYRROLIDINYL)ETHYL]PHENYL}-3-( TRIFLUOROMETHYL)-4,5,6,7-TETRAHYDRO-1H-INDAZOLE, GLUTAMATE RECEPTOR 2, GLUTAMIC ACID, ... | | Authors: | Ward, S.E, Harries, M, Aldegheri, L, Austin, N.E, Ballantine, S, Ballini, E, Bradley, D.M, Bax, B.D, Clarke, B.P, Harris, A.J, Harrison, S.A, Melarange, R.A, Mookherjee, C, Mosley, J, DalNegro, G, Oliosi, B, Smith, K.J, Thewlis, K.M, Woollard, P.M, Yusaf, S.P. | | Deposit date: | 2010-11-10 | | Release date: | 2011-04-06 | | Last modified: | 2011-09-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Integration of Lead Optimization with Crystallography for a Membrane-Bound Ion Channel Target: Discovery of a New Class of Ampa Receptor Positive Allosteric Modulators.
J.Med.Chem., 54, 2011
|
|
7OGP
 
 | | Structure of the apo-state of the bacteriophage PhiKZ non-virion RNA polymerase - class including clamp | | Descriptor: | DNA-directed RNA polymerase, PHIKZ055, PHIKZ068, ... | | Authors: | de Martin Garrido, N, Lai Wan Loong, Y.T.E, Yakunina, M, Aylett, C.H.S. | | Deposit date: | 2021-05-07 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the bacteriophage PhiKZ non-virion RNA polymerase.
Nucleic Acids Res., 49, 2021
|
|
8DFT
 
 | | Cryo-EM structure of conjugative pili from Pyrobaculum calidifontis | | Descriptor: | Pilin protein, [(2~{S},7~{S},11~{S},15~{S},19~{R},22~{R},26~{S},30~{R},34~{R},38~{S},43~{S},47~{S},51~{S},55~{R},58~{R},62~{S},66~{R},70~{R})-38-(hydroxymethyl)-7,11,15,19,22,26,30,34,43,47,51,55,58,62,66,70-hexadecamethyl-1,4,37,40-tetraoxacyclodoheptacont-2-yl]methanol | | Authors: | Beltran, L.C, Egelman, E.H. | | Deposit date: | 2022-06-22 | | Release date: | 2023-03-22 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Archaeal DNA-import apparatus is homologous to bacterial conjugation machinery.
Nat Commun, 14, 2023
|
|
7OGR
 
 | | Structure of the apo-state of the bacteriophage PhiKZ non-virion RNA polymerase | | Descriptor: | DNA-directed RNA polymerase, PHIKZ055, PHIKZ068, ... | | Authors: | de Martin Garrido, N, Lai Wan Loong, Y.T.E, Yakunina, M, Aylett, C.H.S. | | Deposit date: | 2021-05-07 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of the bacteriophage PhiKZ non-virion RNA polymerase.
Nucleic Acids Res., 49, 2021
|
|
8HQR
 
 | |
2XX8
 
 | | Crystal structure of N,N-dimethyl-4-(3-(trifluoromethyl)-4,5,6,7- tetrahydro-1H-indazol-1-yl)benzamide in complex with the ligand binding domain of the Rat GluA2 receptor and glutamate at 2.2A resolution. | | Descriptor: | GLUTAMATE RECEPTOR 2, GLUTAMIC ACID, N,N-DIMETHYL-4-[3-(TRIFLUOROMETHYL)-4,5,6,7-TETRAHYDRO-1H-INDAZOL-1-YL]BENZAMIDE, ... | | Authors: | Ward, S.E, Harries, M, Aldegheri, L, Austin, N.E, Ballantine, S, Ballini, E, Bradley, D.M, Bax, B.D, Clarke, B.P, Harris, A.J, Harrison, S.A, Melarange, R.A, Mookherjee, C, Mosley, J, DalNegro, G, Oliosi, B, Smith, K.J, Thewlis, K.M, Woollard, P.M, Yusaf, S.P. | | Deposit date: | 2010-11-09 | | Release date: | 2011-04-27 | | Last modified: | 2011-09-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Integration of Lead Optimization with Crystallography for a Membrane-Bound Ion Channel Target: Discovery of a New Class of Ampa Receptor Positive Allosteric Modulators.
J.Med.Chem., 54, 2011
|
|
