4YEC
 
 | |
6PM4
 
 | | CryoEM structure of zebra fish alpha-1 glycine receptor bound with Glycine in SMA, super-open state | | Descriptor: | GLYCINE, Glycine receptor subunit alphaZ1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yu, J, Zhu, H, Gouaux, E. | | Deposit date: | 2019-07-01 | | Release date: | 2021-02-10 | | Last modified: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Mechanism of gating and partial agonist action in the glycine receptor.
Cell, 184, 2021
|
|
6PLP
 
 | | CryoEM structure of zebra fish alpha-1 glycine receptor YGF mutant bound with GABA in SMA, desensitized state | | Descriptor: | GAMMA-AMINO-BUTANOIC ACID, Glycine receptor subunit alphaZ1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yu, J, Zhu, H, Gouaux, E. | | Deposit date: | 2019-07-01 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mechanism of gating and partial agonist action in the glycine receptor.
Cell, 184, 2021
|
|
6PM2
 
 | | CryoEM structure of zebra fish alpha-1 glycine receptor bound with Taurine in SMA, open state | | Descriptor: | 2-AMINOETHANESULFONIC ACID, Glycine receptor subunit alphaZ1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yu, J, Zhu, H, Gouaux, E. | | Deposit date: | 2019-07-01 | | Release date: | 2021-02-10 | | Last modified: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanism of gating and partial agonist action in the glycine receptor.
Cell, 184, 2021
|
|
6PM6
 
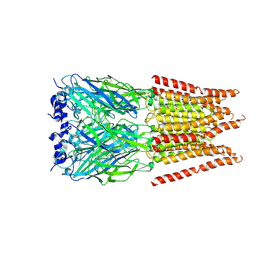 | | CryoEM structure of zebra fish alpha-1 glycine receptor bound with Glycine in SMA, open state | | Descriptor: | GLYCINE, Glycine receptor subunit alphaZ1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yu, J, Zhu, H, Gouaux, E. | | Deposit date: | 2019-07-01 | | Release date: | 2021-02-10 | | Last modified: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mechanism of gating and partial agonist action in the glycine receptor.
Cell, 184, 2021
|
|
6PLW
 
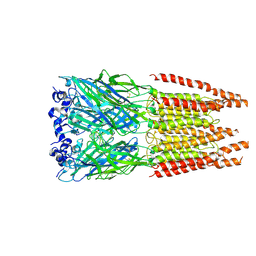 | | CryoEM structure of zebra fish alpha-1 glycine receptor bound with GABA in SMA, super-open state | | Descriptor: | GAMMA-AMINO-BUTANOIC ACID, Glycine receptor subunit alphaZ1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yu, J, Zhu, H, Gouaux, E. | | Deposit date: | 2019-07-01 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanism of gating and partial agonist action in the glycine receptor.
Cell, 184, 2021
|
|
6YSA
 
 | | Crystal structure of Arabidopsis thaliana legumain isoform beta in zymogen state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, SULFATE ION, ... | | Authors: | Dall, E, Zauner, F.B, Brandstetter, H. | | Deposit date: | 2020-04-21 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural and functional studies ofArabidopsis thalianalegumain beta reveal isoform specific mechanisms of activation and substrate recognition.
J.Biol.Chem., 295, 2020
|
|
3LEU
 
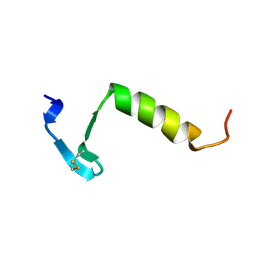 | | HIGH RESOLUTION 1H NMR STUDY OF LEUCOCIN A IN DODECYLPHOSPHOCHOLINE MICELLES, 19 STRUCTURES (1:40 RATIO OF LEUCOCIN A:DPC) (0.1% TFA) | | Descriptor: | LEUCOCIN A | | Authors: | Gallagher, N.L.F, Sailer, M, Niemczura, W.P, Nakashima, T.T, Stiles, M.E, Vederas, J.C. | | Deposit date: | 1997-05-20 | | Release date: | 1997-11-26 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of leucocin A in trifluoroethanol and dodecylphosphocholine micelles: spatial location of residues critical for biological activity in type IIa bacteriocins from lactic acid bacteria.
Biochemistry, 36, 1997
|
|
6PQT
 
 | |
5MU8
 
 | | HUMAN TNF-ALPHA IN COMPLEX WITH JNJ525 | | Descriptor: | Tumor necrosis factor, ~{N}4-(phenylmethyl)-~{N}4-[2-[3-(2-piperazin-1-ylpyrimidin-5-yl)phenyl]phenyl]pyrimidine-2,4-diamine | | Authors: | Blevitt, J.M, Hack, M.D, Herman, K.L, Jackson, P.F, Krawczuk, P.J, Lebsack, A.D, Liu, A.X, Mirzadegan, T, Nelen, M.I, Patrick, A.P, Steinbacher, S, Milla, M.E, Lumb, K.J. | | Deposit date: | 2017-01-12 | | Release date: | 2017-03-29 | | Last modified: | 2019-02-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis of Small-Molecule Aggregate Induced Inhibition of a Protein-Protein Interaction.
J. Med. Chem., 60, 2017
|
|
1B9Z
 
 | | BACILLUS CEREUS BETA-AMYLASE COMPLEXED WITH MALTOSE | | Descriptor: | ACETATE ION, CALCIUM ION, PROTEIN (BETA-AMYLASE), ... | | Authors: | Mikami, B, Adachi, M, Kage, T, Sarikaya, E, Nanmori, T, Shinke, R, Utsumi, S. | | Deposit date: | 1999-03-06 | | Release date: | 1999-03-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of raw starch-digesting Bacillus cereus beta-amylase complexed with maltose.
Biochemistry, 38, 1999
|
|
6EUU
 
 | | The Transcriptional Regulator PrfA from Listeria Monocytogenes in complex with a ring-fused 2-pyridone (KSK29) | | Descriptor: | (3~{R})-8-cyclopropyl-5-oxidanylidene-7-(quinolin-3-ylmethyl)-2,3-dihydro-[1,3]thiazolo[3,2-a]pyridine-3-carboxylic acid, ISOPROPYL ALCOHOL, Listeriolysin positive regulatory factor A, ... | | Authors: | Begum, A, Hall, M, Grundstrom, C, Kulen, M, Lindgren, M, Johansson, J, Almqvist, F, Sauer, U.H, Sauer-Eriksson, A.E. | | Deposit date: | 2017-10-31 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Based Design of Inhibitors Targeting PrfA, the Master Virulence Regulator of Listeria monocytogenes.
J. Med. Chem., 61, 2018
|
|
8BCS
 
 | | X-ray crystal structure of a de novo designed helix-loop-helix homodimer in an anti arrangement, CC-HP1.0 | | Descriptor: | ACETATE ION, CC-HP1.0 | | Authors: | Edgell, C.L, Mylemans, B, Naudin, E.A, Smith, A.J, Savery, N.J, Woolfson, D.N. | | Deposit date: | 2022-10-17 | | Release date: | 2023-06-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design and Selection of Heterodimerizing Helical Hairpins for Synthetic Biology.
Acs Synth Biol, 12, 2023
|
|
6YCC
 
 | | Structure the ananain protease from Ananas comosus covalently bound to the E64 inhibitor | | Descriptor: | Ananain, GLYCEROL, N-[N-[1-HYDROXYCARBOXYETHYL-CARBONYL]LEUCYLAMINO-BUTYL]-GUANIDINE, ... | | Authors: | Azarkan, M, Charlier, P, Herman, R, Delbrassine, F, Sauvage, E, M Rabet, N, Calvo Esposito, R, Kerff, F. | | Deposit date: | 2020-03-18 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of the free and inhibitors-bound forms of bromelain and ananain from Ananas comosus stem and in vitro study of their cytotoxicity.
Sci Rep, 10, 2020
|
|
1B94
 
 | | RESTRICTION ENDONUCLEASE ECORV WITH CALCIUM | | Descriptor: | CALCIUM ION, DNA (5'-D(*AP*AP*AP*GP*AP*TP*AP*TP*CP*TP*T)-3'), RESTRICTION ENDONUCLEASE ECORV | | Authors: | Thomas, M.P, Halford, S.E, Brady, R.L. | | Deposit date: | 1999-02-19 | | Release date: | 1999-02-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of a mutational hot-spot in the EcoRV restriction endonuclease: a catalytic role for a main chain carbonyl group.
Nucleic Acids Res., 27, 1999
|
|
5UI4
 
 | | Structure of NME1 covalently conjugated to imidazole fluorosulfate | | Descriptor: | 4-[4-(3-methoxyphenyl)-1-(prop-2-yn-1-yl)-1H-imidazol-5-yl]phenyl sulfurofluoridate, Nucleoside diphosphate kinase A | | Authors: | Mortenson, D.E, Brighty, G.J, Wilson, I.A, Kelly, J.W. | | Deposit date: | 2017-01-12 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | "Inverse Drug Discovery" Strategy To Identify Proteins That Are Targeted by Latent Electrophiles As Exemplified by Aryl Fluorosulfates.
J. Am. Chem. Soc., 140, 2018
|
|
2JDM
 
 | | Mutant (S22A) of Pseudomonas aeruginosa lectin II (PA-IIL) complexed with methyl-a-L-fucopyranoside | | Descriptor: | CALCIUM ION, FUCOSE-BINDING LECTIN PA-IIL, SULFATE ION, ... | | Authors: | Adam, J, Pokorna, M, Sabin, C, Mitchell, E.P, Imberty, A, Wimmerova, M. | | Deposit date: | 2007-01-11 | | Release date: | 2007-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Engineering of Pa-Iil Lectin from Pseudomonas Aeruginosa - Unravelling the Role of the Specificity Loop for Sugar Preference.
Bmc Struct.Biol., 7, 2007
|
|
1BCC
 
 | | CYTOCHROME BC1 COMPLEX FROM CHICKEN | | Descriptor: | 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine, FE2/S2 (INORGANIC) CLUSTER, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Zhang, Z, Huang, L, Shulmeister, V.M, Chi, Y.-I, Kim, K.K, Hung, L.-W, Crofts, A.R, Berry, E.A, Kim, S.-H. | | Deposit date: | 1998-03-23 | | Release date: | 1998-08-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Electron transfer by domain movement in cytochrome bc1.
Nature, 392, 1998
|
|
6EXR
 
 | | CHEMOTAXIS PROTEIN CHEY FROM Pyrococcus horikoshiI | | Descriptor: | 120aa long hypothetical chemotaxis protein (CheY) | | Authors: | Paithankar, K.S, Enderle, M.E, Wirthensohn, D, Grininger, M, Oesterhelt, D. | | Deposit date: | 2017-11-09 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structure of the archaeal chemotaxis protein CheY in a domain-swapped dimeric conformation.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
7Z2P
 
 | | Tubulin-nocodazole complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Prota, A.E, Muehlethaler, T, Cavalli, A, Steinmetz, M.O. | | Deposit date: | 2022-02-28 | | Release date: | 2022-08-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Novel fragment-derived colchicine-site binders as microtubule-destabilizing agents.
Eur.J.Med.Chem., 241, 2022
|
|
6B8J
 
 | |
7Z2N
 
 | | Tubulin-18-complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Muehlethaler, T, Prota, A.E, Cavalli, A, Steinmetz, M.O. | | Deposit date: | 2022-02-28 | | Release date: | 2022-08-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Novel fragment-derived colchicine-site binders as microtubule-destabilizing agents.
Eur.J.Med.Chem., 241, 2022
|
|
8BQF
 
 | | Adenylate Kinase L107I MUTANT | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE | | Authors: | Scheerer, D, Adkar, B.V, Bhattacharyya, S, Levy, D, Iljina, M, Iljina, I, Dym, O, Haran, G, Shakhnovich, E.I. | | Deposit date: | 2022-11-21 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Allosteric communication between ligand binding domains modulates substrate inhibition in adenylate kinase.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
1BAZ
 
 | |
7S51
 
 | | Structure of C208A Sortase A from Streptococcus pyogenes bound to LPATA peptide | | Descriptor: | LEU-PRO-ALA-THR-ALA, Sortase | | Authors: | Johnson, D.A, Svendsen, J.E, Antos, J.M, Amacher, J.F. | | Deposit date: | 2021-09-09 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structures of Streptococcus pyogenes class A sortase in complex with substrate and product mimics provide key details of target recognition.
J.Biol.Chem., 298, 2022
|
|
