8V0R
 
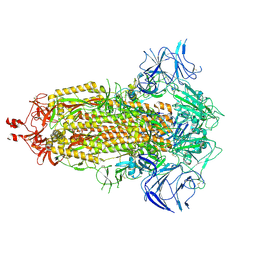 | | SARS-CoV-2 Omicron-XBB.1.5 3-RBD down Spike Protein Trimer 1 (S-GSAS-Omicron-XBB.1.5) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, Q.E, Acharya, P. | | Deposit date: | 2023-11-17 | | Release date: | 2024-06-12 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | SARS-CoV-2 Omicron XBB lineage spike structures, conformations, antigenicity, and receptor recognition.
Mol.Cell, 84, 2024
|
|
4Z6I
 
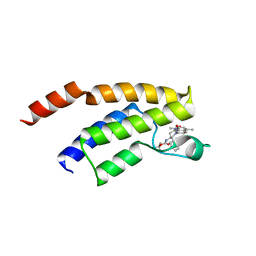 | | Crystal structure of BRD9 bromodomain in complex with a substituted valerolactam quinolone ligand | | Descriptor: | Bromodomain-containing protein 9, tert-butyl [(2R,3S)-1-(1,4-dimethyl-2-oxo-1,2-dihydroquinolin-7-yl)-6-oxo-2-phenylpiperidin-3-yl]carbamate | | Authors: | Tallant, C, Structural Genomics Consortium (SGC), Clark, P.G.K, Vieira, L.C.C, Newman, J.A, Krojer, T, Nunez-Alonso, G, Picaud, S, Fedorov, O, Dixon, D.J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brennan, P.E, Knapp, S. | | Deposit date: | 2015-04-05 | | Release date: | 2015-05-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | LP99: Discovery and Synthesis of the First Selective BRD7/9 Bromodomain Inhibitor.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4Z6P
 
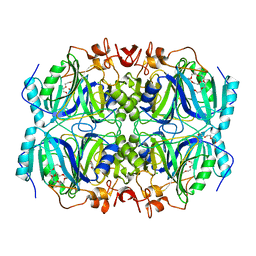 | | Structure of H200Q variant of Homoprotocatechuate 2,3-Dioxygenase from B.fuscum in complex with HPCA at 1.75 Ang resolution | | Descriptor: | 2-(3,4-DIHYDROXYPHENYL)ACETIC ACID, CHLORIDE ION, FE (II) ION, ... | | Authors: | Kovaleva, E.G, Lipscomb, J.D. | | Deposit date: | 2015-04-06 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for Substrate and Oxygen Activation in Homoprotocatechuate 2,3-Dioxygenase: Roles of Conserved Active Site Histidine 200.
Biochemistry, 54, 2015
|
|
6V9H
 
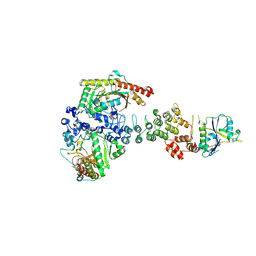 | | Ankyrin repeat and SOCS-box protein 9 (ASB9), ElonginB (ELOB), and ElonginC (ELOC) bound to its substrate Brain-type Creatine Kinase (CKB) | | Descriptor: | Ankyrin repeat and SOCS box protein 9, Creatine kinase B-type, Elongin-B, ... | | Authors: | Komives, E.A, Lumpkin, R.J, Baker, R.W, Leschziner, A.E. | | Deposit date: | 2019-12-13 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure and dynamics of the ASB9 CUL-RING E3 Ligase.
Nat Commun, 11, 2020
|
|
4Z7P
 
 | | X-ray structure of racemic ShK Q16K toxin | | Descriptor: | Potassium channel toxin kappa-stichotoxin-She1a, SULFATE ION | | Authors: | Sickmier, E.A. | | Deposit date: | 2015-04-07 | | Release date: | 2015-09-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Pharmaceutical Optimization of Peptide Toxins for Ion Channel Targets: Potent, Selective, and Long-Lived Antagonists of Kv1.3.
J.Med.Chem., 58, 2015
|
|
8OG1
 
 | | Exostosin-like 3 apo enzyme | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Exostosin-like 3, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Sammon, D, Hohenester, E. | | Deposit date: | 2023-03-17 | | Release date: | 2023-07-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Molecular mechanism of decision-making in glycosaminoglycan biosynthesis.
Nat Commun, 14, 2023
|
|
8C9K
 
 | |
7NLZ
 
 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with 5-Methoxy-6-(trifluoromethyl)indole. | | Descriptor: | 5-methoxy-6-(trifluoromethyl)-1~{H}-indole, Acetylglutamate kinase, SULFATE ION | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-22 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.163 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
8V0Q
 
 | | SARS-CoV-2 Omicron-XBB.1.16 3-RBD down Spike Protein Trimer 3 (S-GSAS-Omicron-XBB.1.16) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, Q.E, Acharya, P. | | Deposit date: | 2023-11-17 | | Release date: | 2024-06-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | SARS-CoV-2 Omicron XBB lineage spike structures, conformations, antigenicity, and receptor recognition.
Mol.Cell, 84, 2024
|
|
6FRO
 
 | | Crystal structure of Hen Egg-White Lysozyme co-crystallized in presence of 100 mM Tb-Xo4 and 100 mM potassium iodide. | | Descriptor: | IODIDE ION, Lysozyme C, SODIUM ION, ... | | Authors: | Engilberge, S, Riobe, F, Di Pietro, S, Girard, E, Dumont, E, Maury, O. | | Deposit date: | 2018-02-16 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Unveiling the Binding Modes of the Crystallophore, a Terbium-based Nucleating and Phasing Molecular Agent for Protein Crystallography.
Chemistry, 24, 2018
|
|
8V0U
 
 | | SARS-CoV-2 Omicron-XBB.1.5 3-RBD down Spike Protein Trimer 4 (S-GSAS-Omicron-XBB.1.5) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, Q.E, Acharya, P. | | Deposit date: | 2023-11-17 | | Release date: | 2024-06-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | SARS-CoV-2 Omicron XBB lineage spike structures, conformations, antigenicity, and receptor recognition.
Mol.Cell, 84, 2024
|
|
4ZET
 
 | | Blood dendritic cell antigen 2 (BDCA-2) complexed with GalGlcNAcMan | | Descriptor: | C-type lectin domain family 4 member C, CALCIUM ION, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose | | Authors: | Jegouzo, S.A.F, Feinberg, H, Dungarwalla, T, Drickamer, K, Weis, W.I, Taylor, M.E. | | Deposit date: | 2015-04-20 | | Release date: | 2015-05-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A Novel Mechanism for Binding of Galactose-terminated Glycans by the C-type Carbohydrate Recognition Domain in Blood Dendritic Cell Antigen 2.
J.Biol.Chem., 290, 2015
|
|
1LH7
 
 | | X-RAY STRUCTURAL INVESTIGATION OF LEGHEMOGLOBIN. VI. STRUCTURE OF ACETATE-FERRILEGHEMOGLOBIN AT A RESOLUTION OF 2.0 ANGSTROMS (RUSSIAN) | | Descriptor: | LEGHEMOGLOBIN (NITROSOBENZENE), NITROSOBENZENE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Vainshtein, B.K, Harutyunyan, E.H, Kuranova, I.P, Borisov, V.V, Sosfenov, N.I, Pavlovsky, A.G, Grebenko, A.I, Konareva, N.V. | | Deposit date: | 1982-04-23 | | Release date: | 1983-01-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-Ray Structural Investigation of Leghemoglobin. Vi. Structure of Acetate-Ferrileghemoglobin at a Resolution of 2.0 Angstroms (Russian)
Kristallografiya, 25, 1980
|
|
6VC4
 
 | | Peanut lectin complexed with S-beta-D-Thiogalactopyranosyl beta-D-glucopyranoside derivative (STGD) | | Descriptor: | (2R,3R,4S,5R,6S)-2-(hydroxymethyl)-6-{[(2S,3R,4S,5S,6S)-3,4,5-trihydroxy-6-({[(1-{[(2R,3S,4S,5R,6S)-3,4,5-trihydroxy-6-methoxytetrahydro-2H-pyran-2-yl]methyl}-1H-1,2,3-triazol-4-yl)methyl]sulfanyl}methyl)tetrahydro-2H-pyran-2-yl]sulfanyl}tetrahydro-2H-pyran-3,4,5-triol (non-preferred name), CALCIUM ION, Galactose-binding lectin, ... | | Authors: | Otero, L.H, Primo, E.D, Cagnoni, A.J, Cano, M.E, Klinke, S, Goldbaum, F.A, Uhrig, M.L. | | Deposit date: | 2019-12-20 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of peanut lectin in the presence of synthetic beta-N- and beta-S-galactosides disclose evidence for the recognition of different glycomimetic ligands.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6NLE
 
 | | X-ray structure of LeuT with V269 deletion | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Navratna, V, Yang, D, Gouaux, E. | | Deposit date: | 2019-01-08 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.615 Å) | | Cite: | Structural, functional, and behavioral insights of dopamine dysfunction revealed by a deletion inSLC6A3.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
8CGC
 
 | | Structure of CSF1R in complex with a pyrollopyrimidine (compound 23) | | Descriptor: | (2S)-2-hydroxybutanedioic acid, GLYCEROL, Macrophage colony-stimulating factor 1 receptor, ... | | Authors: | Aarhus, T.I, Bjornstad, F, Wolowczyk, C, Larsen, K.U, Rognstad, L, Leithaug, T, Unger, A, Habenberger, P, Wolff, A, Bjorkoy, G, Pridans, C, Eickhoff, J, Klebl, B, Hoff, B.H, Sundby, E. | | Deposit date: | 2023-02-03 | | Release date: | 2023-05-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.925 Å) | | Cite: | Synthesis and Development of Highly Selective Pyrrolo[2,3- d ]pyrimidine CSF1R Inhibitors Targeting the Autoinhibited Form.
J.Med.Chem., 66, 2023
|
|
1SUO
 
 | | Structure of mammalian cytochrome P450 2B4 with bound 4-(4-chlorophenyl)imidazole | | Descriptor: | 4-(4-CHLOROPHENYL)IMIDAZOLE, Cytochrome P450 2B4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Scott, E.E, White, M.A, He, Y.A, Johnson, E.F, Stout, C.D, Halpert, J.R. | | Deposit date: | 2004-03-26 | | Release date: | 2004-07-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of mammalian cytochrome P450 2B4 complexed with 4-(4-chlorophenyl)imidazole at 1.9 {angstrom} resolution: Insight into the range of P450 conformations and coordination of redox partner binding.
J.Biol.Chem., 279, 2004
|
|
6NPN
 
 | | Crystal Structure of the Human vaccinia-related kinase bound to a N,N-dipropynyl-dihydropteridine-3-hydroxyindazole inhibitor | | Descriptor: | (7R)-7-methyl-2-[(3-oxo-2,3-dihydro-1H-indazol-6-yl)amino]-5,8-di(prop-2-yn-1-yl)-7,8-dihydropteridin-6(5H)-one, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Counago, R.M, dos Reis, C.V, Takarada, J.E, Azevedo, A, Guimaraes, C, Mascarello, A, Gama, F, Ferreira, M, Massirer, K.B, Arruda, P, Edwards, A.M, Elkins, J.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-01-18 | | Release date: | 2019-02-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Human vaccinia-related kinase bound to a N,N-dipropynyl-dihydropteridine-3-hydroxyindazole inhibitor
To Be Published
|
|
5FKY
 
 | | Structure of a hydrolase bound with an inhibitor | | Descriptor: | (3aR,5R,6S,7R,7aR)-2-amino-5-(hydroxymethyl)-5,6,7,7a-tetrahydro-3aH-pyrano[3,2-d][1,3]thiazole-6,7-diol, GLYCEROL, O-GLCNACASE BT_4395 | | Authors: | Cekic, N, Heinonen, J.E, Stubbs, K.A, Roth, C, McEachern, E.J, Davies, G.J, Vocadlo, D.J. | | Deposit date: | 2015-10-20 | | Release date: | 2016-01-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Analysis of transition state mimicry by tight binding aminothiazoline inhibitors provides insight into catalysis by humanO-GlcNAcase.
Chem Sci, 7, 2016
|
|
5FAQ
 
 | | OXA-48 in complex with FPI-1465 | | Descriptor: | Beta-lactamase, CADMIUM ION, CHLORIDE ION, ... | | Authors: | King, A.M, King, D.T, French, S, Brouillette, E, Asli, A, Alexander, A.N, Vuckovic, M, Maiti, S.N, Parr, T.R, Brown, E.D, Malouin, F, Strynadka, N.C.J, Wright, G.D. | | Deposit date: | 2015-12-11 | | Release date: | 2016-01-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural and Kinetic Characterization of Diazabicyclooctanes as Dual Inhibitors of Both Serine-beta-Lactamases and Penicillin-Binding Proteins.
Acs Chem.Biol., 11, 2016
|
|
8BTK
 
 | | Structure of the TRAP complex with the Sec translocon and a translating ribosome | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | Authors: | Jaskolowski, M, Jomaa, A, Gamerdinger, M, Shrestha, S, Leibundgut, M, Deuerling, E, Ban, N. | | Deposit date: | 2022-11-29 | | Release date: | 2023-05-24 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular basis of the TRAP complex function in ER protein biogenesis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6HAV
 
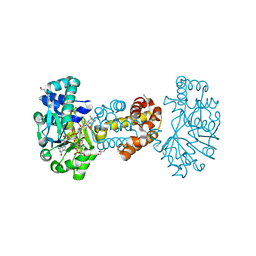 | | Crystal structure of [Fe]-hydrogenase (Hmd) from Methanococcus aeolicus in complex with FeGP and methenyl-tetrahydromethanopterin (close form A) at 1.06 A resolution | | Descriptor: | 1-{4-[(6S,6aR,7R)-3-amino-6,7-dimethyl-1-oxo-1,2,5,6,6a,7-hexahydro-8H-imidazo[1,5-f]pteridin-10-ium-8-yl]phenyl}-1-deoxy-5-O-{5-O-[(S)-{[(1S)-1,3-dicarboxypropyl]oxy}(hydroxy)phosphoryl]-alpha-D-ribofuranosyl}-D-ribitol, 5,10-methenyltetrahydromethanopterin hydrogenase, GLYCEROL, ... | | Authors: | Huang, G, Wagner, T, Wodrich, M.D, Ataka, K, Bill, E, Ermler, U, Hu, X, Shima, S. | | Deposit date: | 2018-08-08 | | Release date: | 2019-08-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | The atomic-resolution crystal structure of activated [Fe]-hydrogenase
Nat Catal, 2019
|
|
4TQC
 
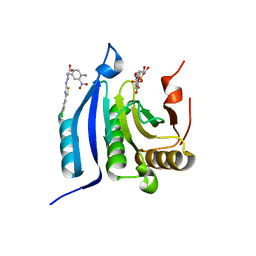 | | The co-complex structure of the translation initiation factor eIF4E with the inhibitor 4EGI-1 reveals an allosteric mechanism for dissociating eIF4G | | Descriptor: | (2S)-3-(4-amino-3-nitrophenyl)-2-{2-[4-(3,4-dichlorophenyl)-1,3-thiazol-2-yl]hydrazinyl}propanoic acid, 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, Eukaryotic translation initiation factor 4E | | Authors: | Papadopoulos, E, Jenni, S, Wagner, G. | | Deposit date: | 2014-06-10 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the eukaryotic translation initiation factor eIF4E in complex with 4EGI-1 reveals an allosteric mechanism for dissociating eIF4G.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4Z6L
 
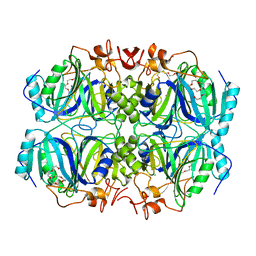 | | Structure of H200E variant of Homoprotocatechuate 2,3-Dioxygenase from B.fuscum at 1.65 Ang resolution | | Descriptor: | CALCIUM ION, CHLORIDE ION, FE (II) ION, ... | | Authors: | Kovaleva, E.G, Lipscomb, J.D. | | Deposit date: | 2015-04-06 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Basis for Substrate and Oxygen Activation in Homoprotocatechuate 2,3-Dioxygenase: Roles of Conserved Active Site Histidine 200.
Biochemistry, 54, 2015
|
|
5F93
 
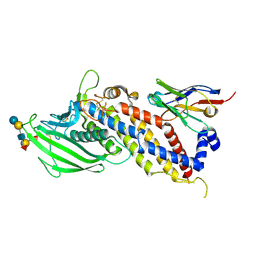 | | Blood group antigen binding adhesin BabA of Helicobacter pylori strain A730 in complex with blood group H Lewis b hexasaccharide | | Descriptor: | Adhesin binding fucosylated histo-blood group antigen,Adhesin,Adhesin binding fucosylated histo-blood group antigen, Nanobody Nb-ER19, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Moonens, K, Gideonsson, P, Subedi, S, Romao, E, Oscarson, S, Muyldermans, S, Boren, T, Remaut, H. | | Deposit date: | 2015-12-09 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural Insights into Polymorphic ABO Glycan Binding by Helicobacter pylori.
Cell Host Microbe, 19, 2016
|
|
