8S9U
 
 | |
8R8Q
 
 | | Lysosomal peptide transporter | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glycosylated lysosomal membrane protein, Major facilitator superfamily domain-containing protein 1 | | Authors: | Jungnickel, K.E.J, Loew, C. | | Deposit date: | 2023-11-29 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.08 Å) | | Cite: | MFSD1 with its accessory subunit GLMP functions as a general dipeptide uniporter in lysosomes.
Nat.Cell Biol., 26, 2024
|
|
4ELK
 
 | | Crystal structure of the Hy19.3 type II NKT TCR | | Descriptor: | ACETATE ION, FORMIC ACID, Hy19.3 TCR alpha chain (mouse variable domain, ... | | Authors: | Girardi, E, Maricic, I, Wang, J, Mac, T.T, Iyer, P, Kumar, V, Zajonc, D.M. | | Deposit date: | 2012-04-10 | | Release date: | 2012-07-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Type II natural killer T cells use features of both innate-like and conventional T cells to recognize sulfatide self antigens.
Nat.Immunol., 13, 2012
|
|
4ZV9
 
 | | 2.00 Angstrom resolution crystal structure of an uncharacterized protein from Escherichia coli O157:H7 str. Sakai | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Halavaty, A.S, Wawrzak, Z, Filippova, E.V, Kiryukhina, O, Endres, M, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-05-18 | | Release date: | 2015-06-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.00 Angstrom resolution crystal structure of an uncharacterized protein from Escherichia coli O157:H7 str. Sakai
To Be Published
|
|
3G1X
 
 | | Crystal structure of the mutant D70G of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with uridine 5'-monophosphate | | Descriptor: | CHLORIDE ION, Orotidine 5'-phosphate decarboxylase, URIDINE-5'-MONOPHOSPHATE | | Authors: | Fedorov, A.A, Fedorov, E.V, Chan, K.K, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2009-01-30 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Mechanism of the orotidine 5'-monophosphate decarboxylase-catalyzed reaction: evidence for substrate destabilization.
Biochemistry, 48, 2009
|
|
2PFJ
 
 | | Crystal Structure of T7 Endo I resolvase in complex with a Holliday Junction | | Descriptor: | 27-MER, CALCIUM ION, Endodeoxyribonuclease 1 | | Authors: | Hadden, J.M, Declais, A.C, Carr, S.B, Lilley, D.M, Phillips, S.E. | | Deposit date: | 2007-04-05 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structural basis of Holliday junction resolution by T7 endonuclease I.
Nature, 449, 2007
|
|
5ACG
 
 | | X-ray Structure of LPMO | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Frandsen, K.E.H, Poulsen, J.N, Tovborg, M, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2015-08-17 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | The molecular basis of polysaccharide cleavage by lytic polysaccharide monooxygenases.
Nat. Chem. Biol., 12, 2016
|
|
3G1D
 
 | | Crystal structure of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with uridine 5'-monophosphate | | Descriptor: | Orotidine 5'-phosphate decarboxylase, URIDINE-5'-MONOPHOSPHATE | | Authors: | Fedorov, A.A, Fedorov, E.V, Chan, K.K, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2009-01-29 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mechanism of the orotidine 5'-monophosphate decarboxylase-catalyzed reaction: evidence for substrate destabilization.
Biochemistry, 48, 2009
|
|
4ZSM
 
 | | BACE crystal structure with bicyclic aminothiazine fragment | | Descriptor: | (4aS,8aR)-4a,5,6,7,8,8a-hexahydro-4H-3,1-benzothiazin-2-amine, Beta-secretase 1, GLYCEROL | | Authors: | Timm, D.E. | | Deposit date: | 2015-05-13 | | Release date: | 2015-06-10 | | Last modified: | 2015-06-17 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Preparation and biological evaluation of conformationally constrained BACE1 inhibitors.
Bioorg.Med.Chem., 23, 2015
|
|
4ZSY
 
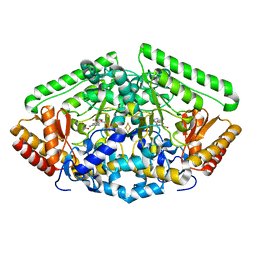 | | Pig Brain GABA-AT inactivated by (Z)-(1S,3S)-3-Amino-4-fluoromethylenyl-1-cyclopentanoic acid. | | Descriptor: | (1S)-4-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]cyclopent-3-ene-1,3-dicarboxylic acid, 4-aminobutyrate aminotransferase, mitochondrial, ... | | Authors: | Wu, R, Lee, H, Le, H.V, Doud, E, Sanishvili, R, Compton, P, Kelleher, N.L, Silverman, R.B, Liu, D. | | Deposit date: | 2015-05-14 | | Release date: | 2015-07-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of Inactivation of GABA Aminotransferase by (E)- and (Z)-(1S,3S)-3-Amino-4-fluoromethylenyl-1-cyclopentanoic Acid.
Acs Chem.Biol., 10, 2015
|
|
5A7O
 
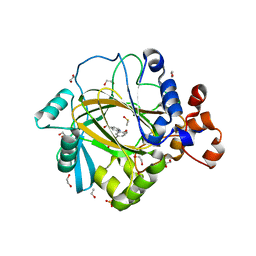 | | Crystal structure of human JMJD2A in complex with compound 42 | | Descriptor: | 1,2-ETHANEDIOL, 2-[5-(2-methoxyethanoylamino)-2-oxidanyl-phenyl]pyridine-4-carboxylic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Nowak, R, Velupillai, S, Krojer, T, Gileadi, C, Johansson, C, Korczynska, M, Le, D.D, Younger, N, Gregori-Puigjane, E, Tumber, A, Iwasa, E, Pollock, S.B, Ortiz Torres, I, Pinkas, D.M, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Shoichet, B.K, Fujimori, D.G, Oppermann, U. | | Deposit date: | 2015-07-09 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Docking and Linking of Fragments to Discover Jumonji Histone Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
4ZSQ
 
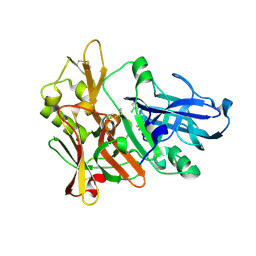 | | BACE crystal structure with tricyclic aminothiazine inhibitor | | Descriptor: | Beta-secretase 1, GLYCEROL, N-[(4S,4aS,6S,8aR)-10-aminohexahydro-3H-4,8a-(epithiomethenoazeno)isochromen-6(1H)-yl]-3-chlorobenzamide | | Authors: | Timm, D.E. | | Deposit date: | 2015-05-13 | | Release date: | 2015-06-10 | | Last modified: | 2015-06-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Preparation and biological evaluation of conformationally constrained BACE1 inhibitors.
Bioorg.Med.Chem., 23, 2015
|
|
5A82
 
 | | Crystal structure of human ATAD2 bromodomain in complex with 4-(3R,4R) -4-(3-methyl-2-oxo-1,2-dihydro-1,7-naphthyridin-8-yl)aminopiperidin-3- yloxymethyl)-1-thiane-1,1-dione | | Descriptor: | 1,2-ETHANEDIOL, 8-[[(3R,4R)-3-[[1,1-bis(oxidanylidene)thian-4-yl]methoxy]piperidin-4-yl]amino]-3-methyl-1H-1,7-naphthyridin-2-one, ATPASE FAMILY AAA DOMAIN-CONTAINING PROTEIN 2, ... | | Authors: | Chung, C, Bamborough, P, Demont, E. | | Deposit date: | 2015-07-11 | | Release date: | 2015-08-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure-Based Optimization of Naphthyridones Into Potent Atad2 Bromodomain Inhibitors.
J.Med.Chem., 58, 2015
|
|
5ACI
 
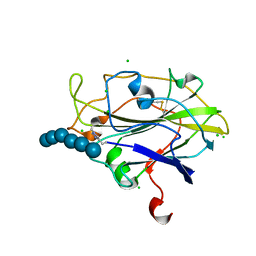 | | X-ray Structure of LPMO | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Frandsen, K.E.H, Poulsen, J.N, Tovborg, M, Johanson, K.S, Lo Leggio, L. | | Deposit date: | 2015-08-17 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The molecular basis of polysaccharide cleavage by lytic polysaccharide monooxygenases.
Nat. Chem. Biol., 12, 2016
|
|
4ZZQ
 
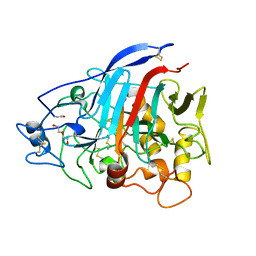 | | Dictyostelium discoideum cellobiohydrolase Cel7A apo structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CELLULOSE 1,4-BETA-CELLOBIOSIDASE, DI(HYDROXYETHYL)ETHER | | Authors: | Momeni, M.H, Hobdey, S.E, Knott, B, Beckham, G.T, Stahlberg, J. | | Deposit date: | 2015-04-13 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biochemical and Structural Characterization of Two Dictyostelium Cellobiohydrolases from the Amoebozoa Kingdom Reveal a High Conservation between Distant Phylogenetic Trees of Life.
Appl.Environ.Microbiol., 82, 2016
|
|
6DRH
 
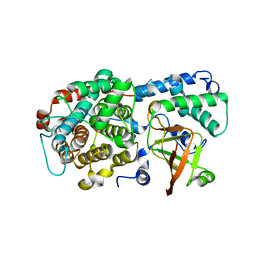 | | ADP-ribosyltransferase toxin/immunity pair | | Descriptor: | ADP-ribosyl-(Dinitrogen reductase) hydrolase, MAGNESIUM ION, PAAR repeat-containing protein, ... | | Authors: | Bosch, D.E, Ting, S, Allaire, M, Mougous, J.D. | | Deposit date: | 2018-06-11 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Bifunctional Immunity Proteins Protect Bacteria against FtsZ-Targeting ADP-Ribosylating Toxins.
Cell, 175, 2018
|
|
1TZ8
 
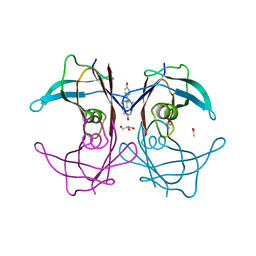 | | The monoclinic crystal structure of transthyretin in complex with diethylstilbestrol | | Descriptor: | ACETATE ION, CARBONATE ION, DIETHYLSTILBESTROL, ... | | Authors: | Morais-de-Sa, E.M, Pereira, P.J.B, Saraiva, M.J, Damas, A.M. | | Deposit date: | 2004-07-09 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The crystal structure of transthyretin in complex with diethylstilbestrol: a promising template for the design of amyloid inhibitors
J.Biol.Chem., 279, 2004
|
|
4N3M
 
 | | Joint neutron/X-ray structure of urate oxidase in complex with 8-azaxanthine | | Descriptor: | 8-AZAXANTHINE, CHLORIDE ION, SODIUM ION, ... | | Authors: | Oksanen, E, Blakeley, M.P, Budayova-Spano, M. | | Deposit date: | 2013-10-07 | | Release date: | 2014-02-05 | | Last modified: | 2018-06-13 | | Method: | NEUTRON DIFFRACTION (1.919 Å), X-RAY DIFFRACTION | | Cite: | The neutron structure of urate oxidase resolves a long-standing mechanistic conundrum and reveals unexpected changes in protonation.
Plos One, 9, 2014
|
|
1U0G
 
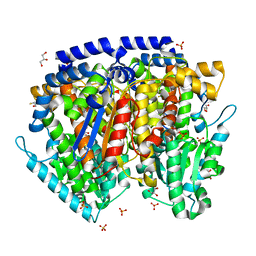 | | Crystal structure of mouse phosphoglucose isomerase in complex with erythrose 4-phosphate | | Descriptor: | BETA-MERCAPTOETHANOL, ERYTHOSE-4-PHOSPHATE, GLYCEROL, ... | | Authors: | Solomons, J.T.G, Zimmerly, E.M, Burns, S, Krishnamurthy, N, Swan, M.K, Krings, S, Muirhead, H, Chirgwin, J, Davies, C. | | Deposit date: | 2004-07-13 | | Release date: | 2004-11-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of mouse phosphoglucose isomerase at 1.6A resolution and its complex with glucose 6-phosphate reveals the catalytic mechanism of sugar ring opening.
J.Mol.Biol., 342, 2004
|
|
1U0F
 
 | | Crystal structure of mouse phosphoglucose isomerase in complex with glucose 6-phosphate | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, BETA-MERCAPTOETHANOL, GLUCOSE-6-PHOSPHATE, ... | | Authors: | Solomons, J.T.G, Zimmerly, E.M, Burns, S, Krishnamurthy, N, Swan, M.K, Krings, S, Muirhead, H, Chirgwin, J, Davies, C. | | Deposit date: | 2004-07-13 | | Release date: | 2004-11-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of mouse phosphoglucose isomerase at 1.6A resolution and its complex with glucose 6-phosphate reveals the catalytic mechanism of sugar ring opening.
J.Mol.Biol., 342, 2004
|
|
4W60
 
 | | The structure of Vaccina virus H7 protein displays A Novel Phosphoinositide binding fold required for membrane biogenesis | | Descriptor: | Late protein H7 | | Authors: | Kolli, S, Meng, X, Wu, X, Shengjuler, D, Cameron, C.E, Xiang, Y, Deng, J. | | Deposit date: | 2014-08-19 | | Release date: | 2014-12-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-function analysis of vaccinia virus h7 protein reveals a novel phosphoinositide binding fold essential for poxvirus replication.
J.Virol., 89, 2015
|
|
6TRE
 
 | | Structure of the RBM3/collar region of the Salmonella flagella MS-ring protein FliF with 32-fold symmetry applied | | Descriptor: | Flagellar M-ring protein | | Authors: | Johnson, S, Fong, Y.H, Deme, J.C, Furlong, E.J, Kuhlen, L, Lea, S.M. | | Deposit date: | 2019-12-18 | | Release date: | 2020-03-18 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the bacterial flagellar rotor MS-ring: a minimum inventory/maximum diversity system.
To Be Published
|
|
1TWS
 
 | | Dihydropteroate Synthetase From Bacillus anthracis | | Descriptor: | DHPS, Dihydropteroate synthase, SULFATE ION | | Authors: | Babaoglu, K, Qi, J, Lee, R.E, White, S.W. | | Deposit date: | 2004-07-01 | | Release date: | 2004-09-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of 7,8-Dihydropteroate Synthase from Bacillus anthracis; Mechanism and Novel Inhibitor Design.
STRUCTURE, 12, 2004
|
|
1TX2
 
 | | Dihydropteroate Synthetase, With Bound Inhibitor MANIC, From Bacillus anthracis | | Descriptor: | 6-METHYLAMINO-5-NITROISOCYTOSINE, DHPS, Dihydropteroate synthase, ... | | Authors: | Babaoglu, K, Qi, J, Lee, R.E, White, S.W. | | Deposit date: | 2004-07-01 | | Release date: | 2004-09-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal Structure of 7,8-Dihydropteroate Synthase from Bacillus anthracis; Mechanism and Novel Inhibitor Design.
STRUCTURE, 12, 2004
|
|
7LWR
 
 | |
