6MKF
 
 | | Crystal structure of penicillin binding protein 5 (PBP5) from Enterococcus faecium in the imipenem-bound form | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carbox ylic acid, SULFATE ION, penicillin binding protein 5 (PBP5) | | Authors: | Moon, T.M, Lee, C, D'Andrea, E.D, Peti, W, Page, R. | | Deposit date: | 2018-09-25 | | Release date: | 2018-10-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structures of penicillin-binding protein 4 (PBP4) and PBP5 fromEnterococciprovide structural insights into beta-lactam resistance.
J. Biol. Chem., 293, 2018
|
|
3M2V
 
 | | Structural Insight into Methyl-Coenzyme M Reductase Chemistry using Coenzyme B Analogues | | Descriptor: | 1,2-ETHANEDIOL, 1-THIOETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Cedervall, P.E, Dey, M, Ragsdale, S.W, Wilmot, C.M. | | Deposit date: | 2010-03-08 | | Release date: | 2010-09-15 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insight into methyl-coenzyme M reductase chemistry using coenzyme B analogues.
Biochemistry, 49, 2010
|
|
7ANT
 
 | | Structure of CYP153A from Polaromonas sp. | | Descriptor: | 1,2-ETHANEDIOL, Cytochrome P450, HEME C | | Authors: | Zukic, E, Rowlinson, B, Sharma, M, Hoffman, S, Hauer, B, Grogan, G. | | Deposit date: | 2020-10-12 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Substrate Anchoring and Flexibility Reduction in CYP153AM.aq Leads to Highly Improved Efficiency toward Octanoic Acid
Acs Catalysis, 11, 2021
|
|
5Y5I
 
 | | Time-resolved SFX structure of cytochrome P450nor: 20 ms after photo-irradiation of caged NO in the presence of NADH (NO-bound state), light data | | Descriptor: | GLYCEROL, NADP nitrous oxide-forming nitric oxide reductase, NITRIC OXIDE, ... | | Authors: | Tosha, T, Nomura, T, Nishida, T, Saeki, N, Okubayashi, K, Yamagiwa, R, Sugahara, M, Nakane, T, Yamashita, K, Hirata, K, Ueno, G, Kimura, T, Hisano, T, Muramoto, K, Sawai, H, Takeda, H, Mizohata, E, Yamashita, A, Kanematsu, Y, Takano, Y, Nango, E, Tanaka, R, Nureki, O, Ikemoto, Y, Murakami, H, Owada, S, Tono, K, Yabashi, M, Yamamoto, M, Ago, H, Iwata, S, Sugimoto, H, Shiro, Y, Kubo, M. | | Deposit date: | 2017-08-09 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
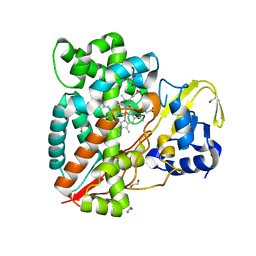
3M4D
 
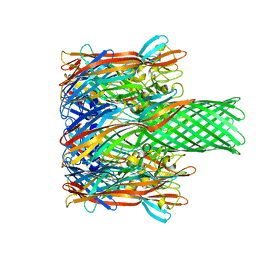 | |
2HMZ
 
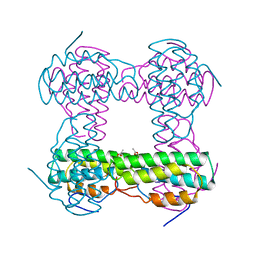 | |
2HMQ
 
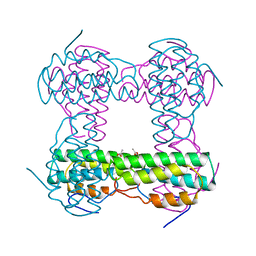 | |
6SSP
 
 | | Structure of the pentameric ligand-gated ion channel ELIC in complex with a NAM nanobody | | Descriptor: | CALCIUM ION, Cys-loop ligand-gated ion channel, NANOBODY 21, ... | | Authors: | Ulens, C, Brams, M, Evans, G.L, Spurny, R, Govaerts, C, Pardon, E, Steyaert, J. | | Deposit date: | 2019-09-09 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Modulation of the Erwinia ligand-gated ion channel (ELIC) and the 5-HT 3 receptor via a common vestibule site.
Elife, 9, 2020
|
|
7YWS
 
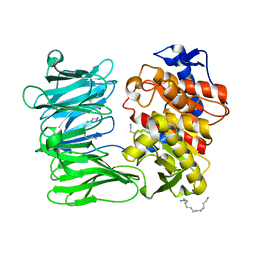 | | Modified oligopeptidase B from S. proteomaculans in intermediate conformation with 3 spermine molecules at 1.7 A resolution | | Descriptor: | Oligopeptidase B, SPERMINE | | Authors: | Petrenko, D.E, Boyko, K.M, Nikolaeva, A.Y, Vlaskina, A.V, Mikhailova, A.G, Timofeev, V.I, Rakitina, T.V. | | Deposit date: | 2022-02-14 | | Release date: | 2023-01-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Elucidation of the Conformational Transition of Oligopeptidase B by an Integrative Approach Based on the Combination of X-ray, SAXS, and Essential Dynamics Sampling Simulation
Crystals, 12, 2022
|
|
7UWH
 
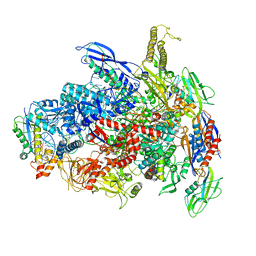 | | CryoEM Structure of E. coli Transcription-Coupled Ribonucleotide Excision Repair (TC-RER) complex bound to ribonucleotide substrate | | Descriptor: | DNA (59-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Hao, Z.T, Grower, M, Bharati, B, Proshkin, S, Epshtein, V, Svetlov, V, Nudler, E, Shamovsky, I. | | Deposit date: | 2022-05-03 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | RNA polymerase drives ribonucleotide excision DNA repair in E. coli.
Cell, 186, 2023
|
|
7ARO
 
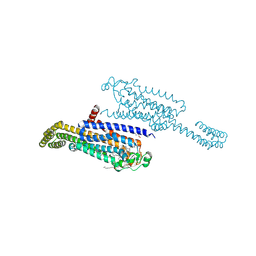 | | Crystal structure of the non-ribose partial agonist LUF5833 bound to the adenosine A2A receptor | | Descriptor: | 2-azanyl-6-(1~{H}-imidazol-2-ylmethylsulfanyl)-4-phenyl-pyridine-3,5-dicarbonitrile, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CHOLESTEROL, ... | | Authors: | Verdon, G, Amelia, T, van Veldhoven, J, Falsini, M, Liu, R, Heitman, L, van Westen, G, Segala, E, Cheng, R, Cooke, R, van der Es, D, Ijzerman, A. | | Deposit date: | 2020-10-25 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.119 Å) | | Cite: | Crystal Structure and Subsequent Ligand Design of a Nonriboside Partial Agonist Bound to the Adenosine A 2A Receptor.
J.Med.Chem., 64, 2021
|
|
7UWE
 
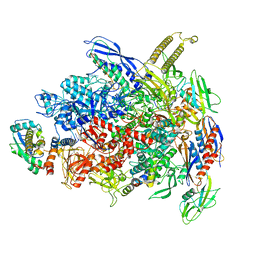 | | CryoEM Structure of E. coli Transcription-Coupled Ribonucleotide Excision Repair (TC-RER) complex | | Descriptor: | DNA (29-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Hao, Z.T, Grower, M, Bharati, B, Proshkin, S, Epshtein, V, Svetlov, V, Nudler, E, Shamovsky, I. | | Deposit date: | 2022-05-03 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | RNA polymerase drives ribonucleotide excision DNA repair in E. coli.
Cell, 186, 2023
|
|
6HTD
 
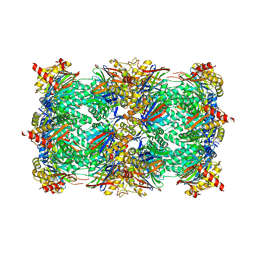 | | Yeast 20S proteasome with human beta2c (S171G) in complex with 4 | | Descriptor: | (2~{S})-~{N}-[(2~{S})-1-[[(2~{S})-1-[4-(aminomethyl)phenyl]-4-methylsulfonyl-butan-2-yl]amino]-1-oxidanylidene-propan-2-yl]-2-[[(2~{S})-2-azido-3-phenyl-propanoyl]amino]-4-methyl-pentanamide, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2018-10-03 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-Based Design of Inhibitors Selective for Human Proteasome beta 2c or beta 2i Subunits.
J.Med.Chem., 62, 2019
|
|
1BH3
 
 | |
6HUU
 
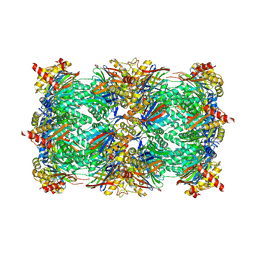 | |
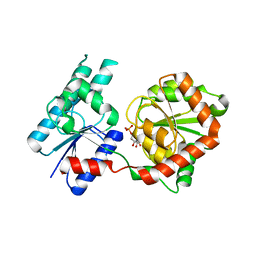
8B5Q
 
 | |
2MBC
 
 | | Solution Structure of human holo-PRL-3 in complex with vanadate | | Descriptor: | Protein tyrosine phosphatase type IVA 3 | | Authors: | Jeong, K, Kang, D, Kim, J, Shin, S, Jin, B, Lee, C, Kim, E, Jeon, Y.H, Kim, Y. | | Deposit date: | 2013-07-29 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and backbone dynamics of vanadate-bound PRL-3: comparison of 15N nuclear magnetic resonance relaxation profiles of free and vanadate-bound PRL-3.
Biochemistry, 53, 2014
|
|
6MKH
 
 | | Crystal structure of pencillin binding protein 4 (PBP4) from Enterococcus faecalis in the imipenem-bound form | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carbox ylic acid, PHOSPHATE ION, pencillin binding protein 4 (PBP4) | | Authors: | D'Andrea, E.D, Moon, T.M, Peti, W, Page, R. | | Deposit date: | 2018-09-25 | | Release date: | 2018-10-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | The structures of penicillin-binding protein 4 (PBP4) and PBP5 fromEnterococciprovide structural insights into beta-lactam resistance.
J. Biol. Chem., 293, 2018
|
|
8B62
 
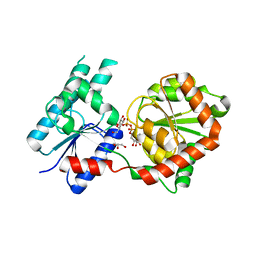 | | Crystal Structure of P. aeruginosa WaaG in complex with UDP-galactose | | Descriptor: | GALACTOSE-URIDINE-5'-DIPHOSPHATE, GLYCEROL, UDP-glucose:(Heptosyl) LPS alpha 1,3-glucosyltransferase WaaG | | Authors: | Scaletti, E, Gustafsson Westergren, R, Stenmark, P. | | Deposit date: | 2022-09-25 | | Release date: | 2023-10-04 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural and functional insights into the Pseudomonas aeruginosa glycosyltransferase WaaG and the implications for lipopolysaccharide biosynthesis.
J.Biol.Chem., 299, 2023
|
|
8B63
 
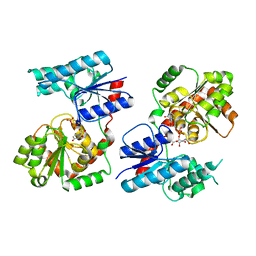 | | Crystal Structure of P. aeruginosa WaaG in complex with UDP-GalNAc | | Descriptor: | ACETATE ION, UDP-glucose:(Heptosyl) LPS alpha 1,3-glucosyltransferase WaaG, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Scaletti, E, Gustafsson Westergren, R, Stenmark, P. | | Deposit date: | 2022-09-25 | | Release date: | 2023-10-04 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional insights into the Pseudomonas aeruginosa glycosyltransferase WaaG and the implications for lipopolysaccharide biosynthesis.
J.Biol.Chem., 299, 2023
|
|
8B5S
 
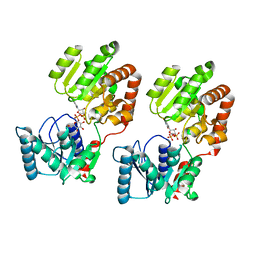 | | Crystal Structure of P. aeruginosa WaaG in complex with UDP-glucose | | Descriptor: | UDP-glucose:(Heptosyl) LPS alpha 1,3-glucosyltransferase WaaG, URIDINE-5'-DIPHOSPHATE, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Scaletti, E, Gustafsson Westergren, R, Stenmark, P. | | Deposit date: | 2022-09-24 | | Release date: | 2023-10-04 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and functional insights into the Pseudomonas aeruginosa glycosyltransferase WaaG and the implications for lipopolysaccharide biosynthesis.
J.Biol.Chem., 299, 2023
|
|
8HK5
 
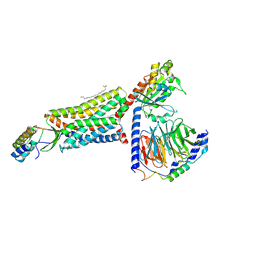 | | C5aR1-Gi-C5a protein complex | | Descriptor: | C5a anaphylatoxin chemotactic receptor 1, Complement C5, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, Y, Liu, W, Xu, Y, Zhuang, Y, Xu, H.E. | | Deposit date: | 2022-11-24 | | Release date: | 2023-05-10 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Revealing the signaling of complement receptors C3aR and C5aR1 by anaphylatoxins.
Nat.Chem.Biol., 19, 2023
|
|
8HK3
 
 | | C3aR-Gi-apo protein complex | | Descriptor: | C3a anaphylatoxin chemotactic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, Y, Liu, W, Xu, Y, Zhuang, Y, Xu, H.E. | | Deposit date: | 2022-11-24 | | Release date: | 2023-05-10 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Revealing the signaling of complement receptors C3aR and C5aR1 by anaphylatoxins.
Nat.Chem.Biol., 19, 2023
|
|
1BE1
 
 | | GLUTAMATE MUTASE (B12-BINDING SUBUNIT), NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | GLUTAMATE MUTASE | | Authors: | Tollinger, M, Konrat, R, Hilbert, B.H, Marsh, E.N.G, Kraeutler, B. | | Deposit date: | 1998-05-19 | | Release date: | 1998-08-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | How a protein prepares for B12 binding: structure and dynamics of the B12-binding subunit of glutamate mutase from Clostridium tetanomorphum
Structure, 6, 1998
|
|
7B27
 
 | |
