4XQW
 
 | | X-ray structure analysis of xylanase-N44E with MES at pH6.0 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Wan, Q, Park, J.M, Riccardi, D.M, Hanson, L.B, Fisher, Z, Smith, J.C, Ostermann, A, Schrader, T, Graham, D.E, Coates, L, Langan, P, Kovalevsky, A.Y. | | Deposit date: | 2015-01-20 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7H1V
 
 | | PanDDA analysis group deposition -- Crystal Structure of ZIKV NS2B-NS3 protease in complex with Z31113727 | | Descriptor: | 1-(pyridin-2-yl)piperidin-4-one, DIMETHYL SULFOXIDE, Serine protease NS3, ... | | Authors: | Ni, X, Godoy, A.S, Marples, P.G, Fairhead, M, Balcomb, B.H, Tomlinson, C.W.E, Koekemoer, L, Aschenbrenner, J.C, Lithgo, R.M, Thompson, W, Wild, C, Williams, E.P, Winokan, M, Chandran, A.V, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-04-03 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | PanDDA analysis group deposition of ZIKV NS2B-NS3 protease
To Be Published
|
|
7H22
 
 | | PanDDA analysis group deposition -- Crystal Structure of ZIKV NS2B-NS3 protease in complex with Z1102357527 | | Descriptor: | DIMETHYL SULFOXIDE, N-[(3R)-6-oxopiperidin-3-yl]-1,3-thiazole-4-carboxamide, Serine protease NS3, ... | | Authors: | Ni, X, Godoy, A.S, Marples, P.G, Fairhead, M, Balcomb, B.H, Tomlinson, C.W.E, Koekemoer, L, Aschenbrenner, J.C, Lithgo, R.M, Thompson, W, Wild, C, Williams, E.P, Winokan, M, Chandran, A.V, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-04-03 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.554 Å) | | Cite: | PanDDA analysis group deposition of ZIKV NS2B-NS3 protease
To Be Published
|
|
3HFU
 
 | | Crystal structure of the ligand binding domain of E. coli CynR with its specific effector azide | | Descriptor: | 1,2-ETHANEDIOL, AZIDE ION, HTH-type transcriptional regulator cynR | | Authors: | Singer, A.U, Evdokimova, E, Kagan, O, Dong, A, Edwards, A.M, Savchenko, A. | | Deposit date: | 2009-05-12 | | Release date: | 2009-06-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the ligand binding domain of E. coli CynR with its specific effector azide
TO BE PUBLISHED
|
|
7H2H
 
 | | PanDDA analysis group deposition -- Crystal Structure of ZIKV NS2B-NS3 protease in complex with Z1575274523 | | Descriptor: | DIMETHYL SULFOXIDE, Serine protease NS3, Serine protease subunit NS2B, ... | | Authors: | Ni, X, Godoy, A.S, Marples, P.G, Fairhead, M, Balcomb, B.H, Tomlinson, C.W.E, Koekemoer, L, Aschenbrenner, J.C, Lithgo, R.M, Thompson, W, Wild, C, Williams, E.P, Winokan, M, Chandran, A.V, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-04-03 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.419 Å) | | Cite: | PanDDA analysis group deposition of ZIKV NS2B-NS3 protease
To Be Published
|
|
7H1L
 
 | | PanDDA analysis group deposition -- Crystal Structure of ZIKV NS2B-NS3 protease in complex with POB0087 | | Descriptor: | (3S)-3-(4-fluorophenyl)piperidine-3-carboxamide, DIMETHYL SULFOXIDE, Serine protease NS3, ... | | Authors: | Ni, X, Godoy, A.S, Marples, P.G, Fairhead, M, Balcomb, B.H, Tomlinson, C.W.E, Koekemoer, L, Aschenbrenner, J.C, Lithgo, R.M, Thompson, W, Wild, C, Williams, E.P, Winokan, M, Chandran, A.V, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-04-03 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.535 Å) | | Cite: | PanDDA analysis group deposition of ZIKV NS2B-NS3 protease
To Be Published
|
|
7GZB
 
 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0000495-001 | | Descriptor: | (2R)-(2,3-dihydro-1-benzofuran-5-yl)[(7H-pyrrolo[2,3-d]pyrimidine-4-carbonyl)amino]acetic acid, Non-structural protein 3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7H1Y
 
 | | PanDDA analysis group deposition -- Crystal Structure of ZIKV NS2B-NS3 protease in complex with Z57493554 | | Descriptor: | 4-(aminomethyl)-1-methylpyridinium, DIMETHYL SULFOXIDE, Serine protease NS3, ... | | Authors: | Ni, X, Godoy, A.S, Marples, P.G, Fairhead, M, Balcomb, B.H, Tomlinson, C.W.E, Koekemoer, L, Aschenbrenner, J.C, Lithgo, R.M, Thompson, W, Wild, C, Williams, E.P, Winokan, M, Chandran, A.V, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-04-03 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.371 Å) | | Cite: | PanDDA analysis group deposition of ZIKV NS2B-NS3 protease
To Be Published
|
|
7RL5
 
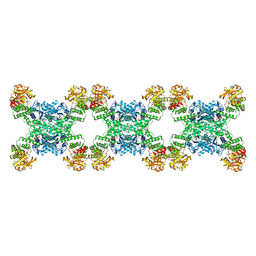 | | Yeast CTP Synthase (URA8) filament bound to CTP at low pH | | Descriptor: | CTP synthase, CYTIDINE-5'-TRIPHOSPHATE | | Authors: | Hansen, J.M, Lynch, E.M, Farrell, D.P, DiMaio, F, Quispe, J, Kollman, J.M. | | Deposit date: | 2021-07-23 | | Release date: | 2021-11-24 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of CTP synthase filaments reveal mechanism of pH-sensitive assembly during budding yeast starvation.
Elife, 10, 2021
|
|
5TIS
 
 | | Room temperature XFEL structure of the native, doubly-illuminated photosystem II complex | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Young, I.D, Ibrahim, M, Chatterjee, R, Gul, S, Fuller, F, Koroidov, S, Brewster, A.S, Tran, R, Alonso-Mori, R, Kroll, T, Michels-Clark, T, Laksmono, H, Sierra, R.G, Stan, C.A, Hussein, R, Zhang, M, Douthit, L, Kubin, M, de Lichtenberg, C, Pham, L.V, Nilsson, H, Cheah, M.H, Shevela, D, Saracini, C, Bean, M.A, Seuffert, I, Sokaras, D, Weng, T.-C, Pastor, E, Weninger, C, Fransson, T, Lassalle, L, Braeuer, P, Aller, P, Docker, P.T, Andi, B, Orville, A.M, Glownia, J.M, Nelson, S, Sikorski, M, Zhu, D, Hunter, M.S, Aquila, A, Koglin, J.E, Robinson, J, Liang, M, Boutet, S, Lyubimov, A.Y, Uervirojnangkoorn, M, Moriarty, N.W, Liebschner, D, Afonine, P.V, Watermann, D.G, Evans, G, Wernet, P, Dobbek, H, Weis, W.I, Brunger, A.T, Zwart, P.H, Adams, P.D, Zouni, A, Messinger, J, Bergmann, U, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2016-10-03 | | Release date: | 2016-11-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.25000381 Å) | | Cite: | Structure of photosystem II and substrate binding at room temperature.
Nature, 540, 2016
|
|
3OPY
 
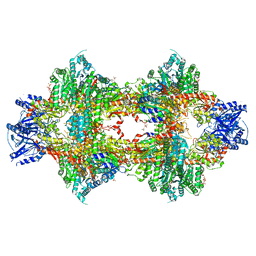 | | Crystal structure of Pichia pastoris phosphofructokinase in the T-state | | Descriptor: | 6-phosphofructo-1-kinase alpha-subunit, 6-phosphofructo-1-kinase beta-subunit, 6-phosphofructo-1-kinase gamma-subunit, ... | | Authors: | Strater, N, Marek, S, Kuettner, E.B, Kloos, M, Keim, A, Bruser, A, Kirchberger, J, Schoneberg, T. | | Deposit date: | 2010-09-02 | | Release date: | 2010-10-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Molecular architecture and structural basis of allosteric regulation of eukaryotic phosphofructokinases.
Faseb J., 25, 2011
|
|
5CKT
 
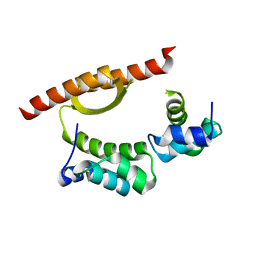 | | Crystal Structure of KorA, a plasmid-encoded, global transcription regulator | | Descriptor: | ACETATE ION, TrfB transcriptional repressor protein | | Authors: | White, S.A, Hyde, E.I, Lovering, A.L. | | Deposit date: | 2015-07-15 | | Release date: | 2016-04-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Flexibility of KorA, a plasmid-encoded, global transcription regulator, in the presence and the absence of its operator.
Nucleic Acids Res., 44, 2016
|
|
7H2F
 
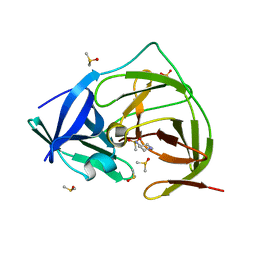 | | PanDDA analysis group deposition -- Crystal Structure of ZIKV NS2B-NS3 protease in complex with Z1491215378 | | Descriptor: | DIMETHYL SULFOXIDE, N-(piperidin-4-yl)methanesulfonamide, SULFATE ION, ... | | Authors: | Ni, X, Godoy, A.S, Marples, P.G, Fairhead, M, Balcomb, B.H, Tomlinson, C.W.E, Koekemoer, L, Aschenbrenner, J.C, Lithgo, R.M, Thompson, W, Wild, C, Williams, E.P, Winokan, M, Chandran, A.V, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-04-03 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.381 Å) | | Cite: | PanDDA analysis group deposition of ZIKV NS2B-NS3 protease
To Be Published
|
|
7H09
 
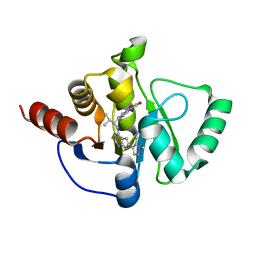 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0011177-001 | | Descriptor: | (4M)-1-methyl-4-(4-{[(1R)-2-methyl-1-(3-oxo-3,4-dihydro-2H-pyrido[3,2-b][1,4]oxazin-6-yl)propyl]amino}-7H-pyrrolo[2,3-d]pyrimidin-6-yl)-1H-pyrazole-5-carbonitrile, Papain-like protease nsp3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7RL0
 
 | | Yeast CTP Synthase (URA8) Filament bound to ATP/UTP at low pH | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CTP synthase, MAGNESIUM ION, ... | | Authors: | Hansen, J.M, Lynch, E.M, Farrell, D.P, DiMaio, F, Quispe, J, Kollman, J.M. | | Deposit date: | 2021-07-22 | | Release date: | 2021-11-24 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures of CTP synthase filaments reveal mechanism of pH-sensitive assembly during budding yeast starvation.
Elife, 10, 2021
|
|
7H0H
 
 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0011204-001 | | Descriptor: | N-ethyl-4-{[(1S)-2-methyl-1-(3-methyl-4-oxo-3,4-dihydroquinazolin-6-yl)propyl]amino}-7H-pyrrolo[2,3-d]pyrimidine-6-carboxamide, Papain-like protease nsp3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7GYY
 
 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0000006-001 | | Descriptor: | 4-[(3S)-3-(1H-1,2,4-triazol-1-yl)pyrrolidin-1-yl]-7H-pyrrolo[2,3-d]pyrimidine, Non-structural protein 3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7D24
 
 | | Hsp90 alpha N-terminal domain in complex with a 4B compund | | Descriptor: | 9-[(3-tert-butyl-1,2-oxazol-5-yl)methyl]-6-chloranyl-purin-2-amine, Heat shock protein HSP 90-alpha | | Authors: | Shin, S.C, Kim, E.E. | | Deposit date: | 2020-09-15 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Basis for Design of New Purine-Based Inhibitors Targeting the Hydrophobic Binding Pocket of Hsp90.
Int J Mol Sci, 21, 2020
|
|
7H0E
 
 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0011153-001 | | Descriptor: | 4-{[(1R)-1-(1,1-dioxo-1,2,3,4-tetrahydro-1lambda~6~-benzothiopyran-7-yl)-2-methylpropyl]amino}-N-methyl-7H-pyrrolo[2,3-d]pyrimidine-5-carboxamide, DI(HYDROXYETHYL)ETHER, Papain-like protease nsp3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7H20
 
 | | PanDDA analysis group deposition -- Crystal Structure of ZIKV NS2B-NS3 protease in complex with Z4605084899 | | Descriptor: | 3-ethenyl-1,2-dimethyl-pyrazole, DIMETHYL SULFOXIDE, SULFATE ION, ... | | Authors: | Ni, X, Godoy, A.S, Marples, P.G, Fairhead, M, Balcomb, B.H, Tomlinson, C.W.E, Koekemoer, L, Aschenbrenner, J.C, Lithgo, R.M, Thompson, W, Wild, C, Williams, E.P, Winokan, M, Chandran, A.V, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-04-03 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | PanDDA analysis group deposition of ZIKV NS2B-NS3 protease
To Be Published
|
|
7D26
 
 | | Hsp90 alpha N-terminal domain in complex with a 8 compund | | Descriptor: | 6-chloranyl-9-[(2-phenyl-1,3-oxazol-5-yl)methyl]purin-2-amine, Heat shock protein HSP 90-alpha | | Authors: | Shin, S.C, Kim, E.E. | | Deposit date: | 2020-09-15 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for Design of New Purine-Based Inhibitors Targeting the Hydrophobic Binding Pocket of Hsp90.
Int J Mol Sci, 21, 2020
|
|
7H2M
 
 | | PanDDA analysis group deposition -- Crystal Structure of ZIKV NS2B-NS3 protease in complex with Z228586974 | | Descriptor: | (3-fluorophenyl)(piperazin-1-yl)methanone, DIMETHYL SULFOXIDE, Serine protease NS3, ... | | Authors: | Ni, X, Godoy, A.S, Marples, P.G, Fairhead, M, Balcomb, B.H, Tomlinson, C.W.E, Koekemoer, L, Aschenbrenner, J.C, Lithgo, R.M, Thompson, W, Wild, C, Williams, E.P, Winokan, M, Chandran, A.V, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-04-03 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.712 Å) | | Cite: | PanDDA analysis group deposition of ZIKV NS2B-NS3 protease
To Be Published
|
|
7D1V
 
 | | Hsp90 alpha N-terminal domain in complex with a 6C compund | | Descriptor: | 6-chloranyl-9-[(3-propan-2-yl-1,2-oxazol-5-yl)methyl]purin-2-amine, Heat shock protein HSP 90-alpha | | Authors: | Shin, S.C, Kim, E.E. | | Deposit date: | 2020-09-15 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.332 Å) | | Cite: | Structural Basis for Design of New Purine-Based Inhibitors Targeting the Hydrophobic Binding Pocket of Hsp90.
Int J Mol Sci, 21, 2020
|
|
7GYZ
 
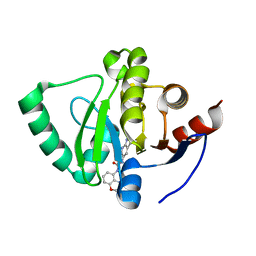 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0000035-001 | | Descriptor: | N-(4-methyl-3,4-dihydro-2H-1,4-benzoxazin-5-yl)-N'-(1H-pyrazolo[3,4-b]pyridin-5-yl)urea, Papain-like protease nsp3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.123 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7D22
 
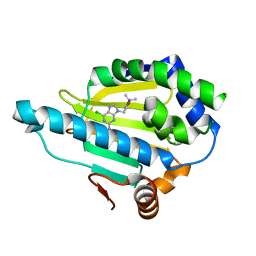 | | Hsp90 alpha N-terminal domain in complex with a 6B compund | | Descriptor: | 9-[(3-tert-butyl-1,2-oxazol-5-yl)methyl]-6-chloranyl-purin-2-amine, Heat shock protein HSP 90-alpha | | Authors: | Shin, S.C, Kim, E.E. | | Deposit date: | 2020-09-15 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis for Design of New Purine-Based Inhibitors Targeting the Hydrophobic Binding Pocket of Hsp90.
Int J Mol Sci, 21, 2020
|
|
