8B41
 
 | |
8B8Q
 
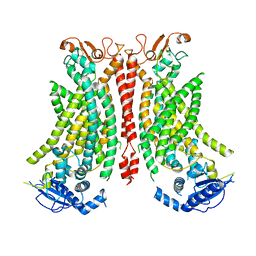 | | Structure of mTMEM16F in lipid Nanodiscs in the presence of Ca2+ | | Descriptor: | Anoctamin-6, CALCIUM ION | | Authors: | Arndt, M, Alvadia, C, Straub, M.S, Clerico-Mosina, V, Paulino, C, Dutzler, R. | | Deposit date: | 2022-10-04 | | Release date: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Structural basis for the activation of the lipid scramblase TMEM16F.
Nat Commun, 13, 2022
|
|
5M8J
 
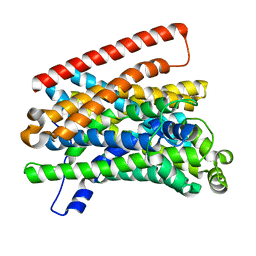 | | Crystal structure of Eremococcus coleocola manganese transporter mutant H236A | | Descriptor: | Divalent metal cation transporter MntH | | Authors: | Manatschal, C, Ehrnstorfer, I.A, Arnold, F.M, Laederach, J, Dutzler, R. | | Deposit date: | 2016-10-29 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural and mechanistic basis of proton-coupled metal ion transport in the SLC11/NRAMP family.
Nat Commun, 8, 2017
|
|
8B42
 
 | |
2HT2
 
 | | Structure of the Escherichia coli ClC chloride channel Y445H mutant and Fab complex | | Descriptor: | BROMIDE ION, Fab fragment, heavy chain, ... | | Authors: | Accardi, A, Lobet, S, Williams, C, Miller, C, Dutzler, R. | | Deposit date: | 2006-07-25 | | Release date: | 2006-09-19 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Synergism Between Halide Binding and Proton Transport in a CLC-type Exchanger.
J.Mol.Biol., 362, 2006
|
|
2EXW
 
 | |
2EXY
 
 | |
2EZ0
 
 | |
7P16
 
 | |
7P5M
 
 | | Cryo-EM structure of human TTYH2 in lipid nanodiscs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Protein tweety homolog 2 | | Authors: | Sukalskaia, A, Straub, M.S, Sawicka, M, Deneka, D, Dutzler, R. | | Deposit date: | 2021-07-14 | | Release date: | 2021-08-11 | | Last modified: | 2021-09-08 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Cryo-EM structures of the TTYH family reveal a novel architecture for lipid interactions.
Nat Commun, 12, 2021
|
|
7P5C
 
 | | Cryo-EM structure of human TTYH3 in Ca2+ and GDN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Protein tweety homolog 3 | | Authors: | Sukalskaia, A, Straub, M.S, Sawicka, M, Deneka, D, Dutzler, R. | | Deposit date: | 2021-07-14 | | Release date: | 2021-08-11 | | Last modified: | 2021-09-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of the TTYH family reveal a novel architecture for lipid interactions.
Nat Commun, 12, 2021
|
|
7P54
 
 | | Cryo-EM structure of human TTYH2 in GDN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Protein tweety homolog 2 | | Authors: | Sukalskaia, A, Straub, M.S, Sawicka, M, Deneka, D, Dutzler, R. | | Deposit date: | 2021-07-14 | | Release date: | 2021-08-11 | | Last modified: | 2021-09-08 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of the TTYH family reveal a novel architecture for lipid interactions.
Nat Commun, 12, 2021
|
|
7P5J
 
 | | Cryo-EM structure of human TTYH1 in GDN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Protein tweety homolog 1 | | Authors: | Sukalskaia, A, Straub, M.S, Sawicka, M, Deneka, D, Dutzler, R. | | Deposit date: | 2021-07-14 | | Release date: | 2021-08-11 | | Last modified: | 2021-09-08 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures of the TTYH family reveal a novel architecture for lipid interactions.
Nat Commun, 12, 2021
|
|
2YN6
 
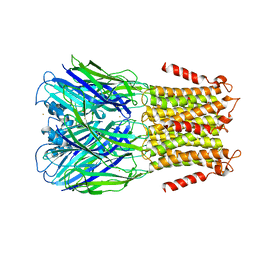 | | Pentameric Ligand-Gated Ion Channel ELIC in Complex with Barium | | Descriptor: | BARIUM ION, PENTAMERIC LIGAND-GATED ION CHANNEL ELIC | | Authors: | Zimmermann, I, Marabelli, A, Bertozzi, C, Sivilotti, L.G, Dutzler, R. | | Deposit date: | 2012-10-12 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Inhibition of the Prokaryotic Pentameric Ligand-Gated Ion Channel Elic by Divalent Cations.
Plos Biol., 10, 2012
|
|
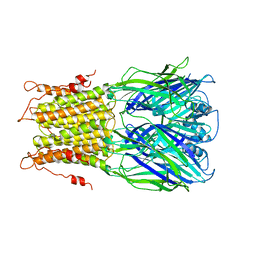
2YKS
 
 | |
2HLF
 
 | | Structure of the Escherichis coli ClC chloride channel Y445E mutant and Fab complex | | Descriptor: | BROMIDE ION, Fab Fragment, Heavy chain, ... | | Authors: | Accardi, A, Lobet, S, Williams, C, Miller, C, Dutzler, R. | | Deposit date: | 2006-07-07 | | Release date: | 2006-09-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Synergism Between Halide Binding and Proton Transport in a CLC-type Exchanger
J.Mol.Biol., 362, 2006
|
|
