2XQ3
 
 | | Pentameric ligand gated ion channel GLIC in complex with Br-lidocaine | | Descriptor: | BROMIDE ION, GLR4197 PROTEIN | | Authors: | Hilf, R.J.C, Bertozzi, C, Zimmermann, I, Reiter, A, Trauner, D, Dutzler, R. | | Deposit date: | 2010-09-01 | | Release date: | 2010-11-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural Basis of Open Channel Block in a Prokaryotic Pentameric Ligand-Gated Ion Channel
Nat.Struct.Mol.Biol., 17, 2010
|
|
6YTX
 
 | |
6YTV
 
 | |
2XQ5
 
 | | Pentameric ligand gated ion channel GLIC in complex with tetraethylarsonium (TEAs) | | Descriptor: | ARSENIC, GLR4197 PROTEIN | | Authors: | Hilf, R.J.C, Bertozzi, C, Zimmermann, I, Reiter, A, Trauner, D, Dutzler, R. | | Deposit date: | 2010-09-01 | | Release date: | 2010-11-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural Basis of Open Channel Block in a Prokaryotic Pentameric Ligand-Gated Ion Channel
Nat.Struct.Mol.Biol., 17, 2010
|
|
5HEJ
 
 | |
5M95
 
 | | STAPHYLOCOCCUS CAPITIS DIVALENT METAL ION TRANSPORTER (DMT) IN COMPLEX WITH MANGANESE | | Descriptor: | CAMELID ANTIBODY FRAGMENT, NANOBODY, Divalent metal cation transporter MntH, ... | | Authors: | Ehrnstorfer, I.A, Geertsma, E.R, Pardon, E, Steyaert, J, Dutzler, R. | | Deposit date: | 2016-10-31 | | Release date: | 2016-11-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal Structure Of A Slc11 (Nramp) Transporter Reveals The Basis For Transition-Metal Ion Transport.
Nat.Struct.Mol.Biol., 21, 2014
|
|
5HEH
 
 | |
5HEW
 
 | | Pentameric ligand-gated ion channel ELIC mutant T28D | | Descriptor: | Gamma-aminobutyric-acid receptor subunit beta-1 | | Authors: | Engeler, S, Dutzler, R. | | Deposit date: | 2016-01-06 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Signal Transduction at the Domain Interface of Prokaryotic Pentameric Ligand-Gated Ion Channels.
Plos Biol., 14, 2016
|
|
5HEO
 
 | |
5HEU
 
 | |
7P14
 
 | | Structure of full-length rXKR9 in complex with a sybody at 3.66A | | Descriptor: | DIUNDECYL PHOSPHATIDYL CHOLINE, O-[(R)-{[(2R)-2,3-bis(octadecanoyloxy)propyl]oxy}(hydroxy)phosphoryl]-L-serine, Sybody, ... | | Authors: | Straub, M.S, Sawicka, M, Dutzler, R. | | Deposit date: | 2021-07-01 | | Release date: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Cryo-EM structures of the caspase activated protein XKR9 involved in apoptotic lipid scrambling.
Elife, 10, 2021
|
|
7P16
 
 | |
7P5M
 
 | | Cryo-EM structure of human TTYH2 in lipid nanodiscs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Protein tweety homolog 2 | | Authors: | Sukalskaia, A, Straub, M.S, Sawicka, M, Deneka, D, Dutzler, R. | | Deposit date: | 2021-07-14 | | Release date: | 2021-08-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Cryo-EM structures of the TTYH family reveal a novel architecture for lipid interactions.
Nat Commun, 12, 2021
|
|
7P5C
 
 | | Cryo-EM structure of human TTYH3 in Ca2+ and GDN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Protein tweety homolog 3 | | Authors: | Sukalskaia, A, Straub, M.S, Sawicka, M, Deneka, D, Dutzler, R. | | Deposit date: | 2021-07-14 | | Release date: | 2021-08-11 | | Last modified: | 2021-09-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of the TTYH family reveal a novel architecture for lipid interactions.
Nat Commun, 12, 2021
|
|
7P54
 
 | | Cryo-EM structure of human TTYH2 in GDN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Protein tweety homolog 2 | | Authors: | Sukalskaia, A, Straub, M.S, Sawicka, M, Deneka, D, Dutzler, R. | | Deposit date: | 2021-07-14 | | Release date: | 2021-08-11 | | Last modified: | 2021-09-08 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of the TTYH family reveal a novel architecture for lipid interactions.
Nat Commun, 12, 2021
|
|
7P5J
 
 | | Cryo-EM structure of human TTYH1 in GDN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Protein tweety homolog 1 | | Authors: | Sukalskaia, A, Straub, M.S, Sawicka, M, Deneka, D, Dutzler, R. | | Deposit date: | 2021-07-14 | | Release date: | 2021-08-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures of the TTYH family reveal a novel architecture for lipid interactions.
Nat Commun, 12, 2021
|
|
5M8K
 
 | | Crystal structure of Eremococcus coleocola manganese transporter mutant E129Q | | Descriptor: | Divalent metal cation transporter MntH | | Authors: | Manatschal, C, Ehrnstorfer, I.A, Arnold, F.M, Laederach, J, Dutzler, R. | | Deposit date: | 2016-10-29 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural and mechanistic basis of proton-coupled metal ion transport in the SLC11/NRAMP family.
Nat Commun, 8, 2017
|
|
5M87
 
 | | Crystal structure of Eremococcus coleocola manganese transporter | | Descriptor: | DECYL-BETA-D-MALTOPYRANOSIDE, Divalent metal cation transporter MntH | | Authors: | Manatschal, C, Ehrnstorfer, I.A, Arnold, F.M, Laederach, J, Dutzler, R. | | Deposit date: | 2016-10-28 | | Release date: | 2017-01-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural and mechanistic basis of proton-coupled metal ion transport in the SLC11/NRAMP family.
Nat Commun, 8, 2017
|
|
5M8A
 
 | | Crystal structure of Eremococcus coleocola manganese transporter mutant E129A | | Descriptor: | Divalent metal cation transporter MntH | | Authors: | Manatschal, C, Ehrnstorfer, I.A, Arnold, F.M, Laederach, J, Dutzler, R. | | Deposit date: | 2016-10-28 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structural and mechanistic basis of proton-coupled metal ion transport in the SLC11/NRAMP family.
Nat Commun, 8, 2017
|
|
5M8J
 
 | | Crystal structure of Eremococcus coleocola manganese transporter mutant H236A | | Descriptor: | Divalent metal cation transporter MntH | | Authors: | Manatschal, C, Ehrnstorfer, I.A, Arnold, F.M, Laederach, J, Dutzler, R. | | Deposit date: | 2016-10-29 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural and mechanistic basis of proton-coupled metal ion transport in the SLC11/NRAMP family.
Nat Commun, 8, 2017
|
|
5NL2
 
 | | cryo-EM structure of the mTMEM16A ion channel at 6.6 A resolution. | | Descriptor: | Anoctamin-1 | | Authors: | Paulino, C, Neldner, Y, Lam, K.M, Kalienkova, V, Brunner, J.D, Schenck, S, Dutzler, R. | | Deposit date: | 2017-04-03 | | Release date: | 2017-06-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structural basis for anion conduction in the calcium-activated chloride channel TMEM16A.
Elife, 6, 2017
|
|
6QMB
 
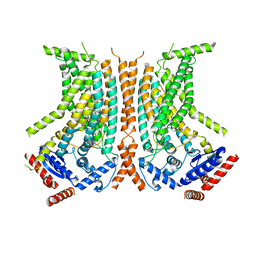 | | Cryo-EM structure of calcium-bound nhTMEM16 lipid scramblase in nanodisc (closed state) | | Descriptor: | CALCIUM ION, Predicted protein | | Authors: | Kalienkova, V, Clerico Mosina, V, Bryner, L, Oostergetel, G.T, Dutzler, R, Paulino, C. | | Deposit date: | 2019-02-01 | | Release date: | 2019-03-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Stepwise activation mechanism of the scramblase nhTMEM16 revealed by cryo-EM.
Elife, 8, 2019
|
|
6QPB
 
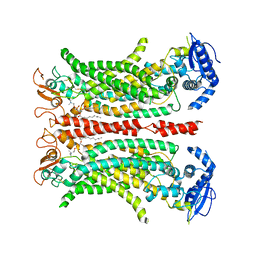 | | Cryo-EM structure of calcium-free mTMEM16F lipid scramblase in digitonin | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Anoctamin-6 | | Authors: | Alvadia, C, Lim, N.K, Clerico Mosina, V, Oostergetel, G.T, Dutzler, R, Paulino, C. | | Deposit date: | 2019-02-13 | | Release date: | 2019-03-06 | | Last modified: | 2019-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures and functional characterization of the murine lipid scramblase TMEM16F.
Elife, 8, 2019
|
|
6QM9
 
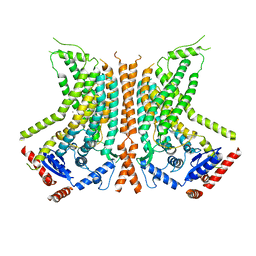 | | Cryo-EM structure of calcium-bound nhTMEM16 lipid scramblase in nanodisc (open state) | | Descriptor: | CALCIUM ION, Predicted protein | | Authors: | Kalienkova, V, Clerico Mosina, V, Bryner, L, Oostergetel, G.T, Dutzler, R, Paulino, C. | | Deposit date: | 2019-02-01 | | Release date: | 2019-03-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Stepwise activation mechanism of the scramblase nhTMEM16 revealed by cryo-EM.
Elife, 8, 2019
|
|
6QM4
 
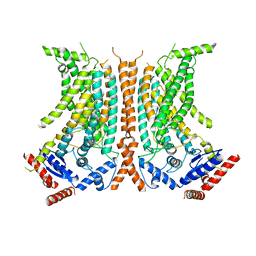 | | Cryo-EM structure of calcium-free nhTMEM16 lipid scramblase in nanodisc | | Descriptor: | Predicted protein | | Authors: | Kalienkova, V, Clerico Mosina, V, Bryner, L, Oostergetel, G.T, Dutzler, R, Paulino, C. | | Deposit date: | 2019-02-01 | | Release date: | 2019-03-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Stepwise activation mechanism of the scramblase nhTMEM16 revealed by cryo-EM.
Elife, 8, 2019
|
|
