6CIB
 
 | |
6CI7
 
 | |
6PEU
 
 | |
2YJY
 
 | | A specific and modular binding code for cytosine recognition in PUF domains | | Descriptor: | 5'-R(*AP*UP*UP*GP*CP*AP*UP*AP*UP*AP)-3', PUMILIO HOMOLOG 1 | | Authors: | Dong, S, Wang, Y, Cassidy-Amstutz, C, Lu, G, Qiu, C, Bigler, R, Jezyk, M, Li, C, Hall, T.M.T, Wang, Z. | | Deposit date: | 2011-05-24 | | Release date: | 2011-06-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Specific and Modular Binding Code for Cytosine Recognition in Pumilio/Fbf (Puf) RNA-Binding Domains.
J.Biol.Chem., 286, 2011
|
|
6WN0
 
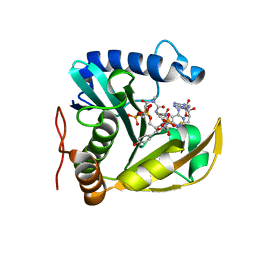 | | The structure of a CoA-dependent acyl-homoserine lactone synthase, RpaI, with the adduct of SAH and p-coumaroyl CoA | | Descriptor: | (2S)-4-({[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}sulfanyl)-2-{[(2E)-3-(cis-4-hydroxycyclohexa-2,5-dien-1-yl)prop-2-enoyl]amino}butanoic acid, 4-coumaroyl-homoserine lactone synthase, COENZYME A | | Authors: | Dong, S.-H, Nair, S.K. | | Deposit date: | 2020-04-22 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Guided Biochemical Analysis of Quorum Signal Synthase Specificities.
Acs Chem.Biol., 15, 2020
|
|
6PE3
 
 | |
6XP8
 
 | |
6OM4
 
 | |
6WNS
 
 | |
8KD5
 
 | | Rpd3S in complex with nucleosome with H3K36MLA modification and 187bp DNA, class2 | | Descriptor: | 187bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Chang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
8KD6
 
 | | Rpd3S in complex with nucleosome with H3K36MLA modification and 187bp DNA, class3 | | Descriptor: | 187bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Zhang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
8KD3
 
 | | Rpd3S in complex with nucleosome with H3K36MLA modification, H3K9Q mutation and 187bp DNA | | Descriptor: | 187bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Zhang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
8KD4
 
 | | Rpd3S in complex with nucleosome with H3K36MLA modification and 187bp DNA, class1 | | Descriptor: | 187bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Zhang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
8KD2
 
 | | Rpd3S in complex with 187bp nucleosome | | Descriptor: | 187bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Zhang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
8KD7
 
 | | Rpd3S in complex with nucleosome with H3K36MLA modification and 167bp DNA | | Descriptor: | 167bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Chang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
8KC7
 
 | | Rpd3S histone deacetylase complex | | Descriptor: | Chromatin modification-related protein EAF3, Histone deacetylase RPD3, Transcriptional regulatory protein RCO1, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Zhang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-06 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
8WBS
 
 | | Crystal structure of cis-Epoxysuccinate Hydrolases KlCESH[L]-D48N complexed with sulfate ions | | Descriptor: | (S)-2-haloacid dehalogenase, CALCIUM ION, SULFATE ION | | Authors: | Dong, S, Xuan, J.S, Feng, Y.G, Cui, Q. | | Deposit date: | 2023-09-10 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Deciphering the stereo-specific catalytic mechanisms of cis-epoxysuccinate hydrolases producing L(+)-tartaric acid.
J.Biol.Chem., 300, 2024
|
|
8WBT
 
 | | Crystal structure of cis-Epoxysuccinate Hydrolases KlCESH[L] mutant D48N complexed with L-TA | | Descriptor: | (S)-2-haloacid dehalogenase, CALCIUM ION, GLYCEROL, ... | | Authors: | Dong, S, Xuan, J.S, Feng, Y.G, Cui, Q. | | Deposit date: | 2023-09-10 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Deciphering the stereo-specific catalytic mechanisms of cis-epoxysuccinate hydrolases producing L(+)-tartaric acid.
J.Biol.Chem., 300, 2024
|
|
5GWE
 
 | | cytochrome P450 CREJ | | Descriptor: | (4-methylphenyl) dihydrogen phosphate, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Dong, S, liu, X, Wang, X, Feng, Y. | | Deposit date: | 2016-09-11 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Selective oxidation of aliphatic C-H bonds in alkylphenols by a chemomimetic biocatalytic system
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8WBO
 
 | | Crystal structure of cis-Epoxysuccinate Hydrolases RhCESH[L] mutant D18N complexed with sulfate ions | | Descriptor: | Epoxide hydrolase, SULFATE ION | | Authors: | Dong, S, Xuan, J.S, Feng, Y.G, Cui, Q. | | Deposit date: | 2023-09-10 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Deciphering the stereo-specific catalytic mechanisms of cis-epoxysuccinate hydrolases producing L(+)-tartaric acid.
J.Biol.Chem., 300, 2024
|
|
8WBR
 
 | | Crystal structure of cis-Epoxysuccinate Hydrolases KlCESH[L] | | Descriptor: | (S)-2-haloacid dehalogenase, CALCIUM ION | | Authors: | Dong, S, Xuan, J.S, Feng, Y.G, Cui, Q. | | Deposit date: | 2023-09-10 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Deciphering the stereo-specific catalytic mechanisms of cis-epoxysuccinate hydrolases producing L(+)-tartaric acid.
J.Biol.Chem., 300, 2024
|
|
8WBK
 
 | | Crystal structure of cis-Epoxysuccinate Hydrolases RhCESH[L] | | Descriptor: | Epoxide hydrolase, SULFATE ION | | Authors: | Dong, S, Xuan, J.S, Feng, Y.G, Cui, Q. | | Deposit date: | 2023-09-10 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Deciphering the stereo-specific catalytic mechanisms of cis-epoxysuccinate hydrolases producing L(+)-tartaric acid.
J.Biol.Chem., 300, 2024
|
|
8WBN
 
 | | Crystal structure of cis-Epoxysuccinate Hydrolases RhCESH[L] mutant D193N | | Descriptor: | Epoxide hydrolase, SULFATE ION | | Authors: | Dong, S, Xuan, J.S, Feng, Y.G, Cui, Q. | | Deposit date: | 2023-09-10 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Deciphering the stereo-specific catalytic mechanisms of cis-epoxysuccinate hydrolases producing L(+)-tartaric acid.
J.Biol.Chem., 300, 2024
|
|
8WBL
 
 | | Crystal structure of cis-Epoxysuccinate Hydrolases RhCESH[L] complexed with sulfate ions | | Descriptor: | Epoxide hydrolase, SULFATE ION | | Authors: | Dong, S, Xuan, J.S, Feng, Y.G, Cui, Q. | | Deposit date: | 2023-09-10 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.941 Å) | | Cite: | Deciphering the stereo-specific catalytic mechanisms of cis-epoxysuccinate hydrolases producing L(+)-tartaric acid.
J.Biol.Chem., 300, 2024
|
|
8WBM
 
 | | Crystal structure of cis-Epoxysuccinate Hydrolases RhCESH[L] mutant D193A complexed with sulfate ions | | Descriptor: | Epoxide hydrolase, SULFATE ION | | Authors: | Dong, S, Xuan, J.S, Feng, Y.G, Cui, Q. | | Deposit date: | 2023-09-10 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Deciphering the stereo-specific catalytic mechanisms of cis-epoxysuccinate hydrolases producing L(+)-tartaric acid.
J.Biol.Chem., 300, 2024
|
|
