6V2H
 
 | | Crystal structure of CDYL2 in complex with H3tK27me3 | | Descriptor: | Chromodomain Y-like protein 2, H3tK27me3, NICKEL (II) ION, ... | | Authors: | Dong, C, Tempel, W, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-11-22 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
2IYD
 
 | | SENP1 covalent complex with SUMO-2 | | Descriptor: | SENTRIN-SPECIFIC PROTEASE 1, SMALL UBIQUITIN-RELATED MODIFIER 2 | | Authors: | Dong, C, Naismith, J.H. | | Deposit date: | 2006-07-14 | | Release date: | 2006-08-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Senp1 Native Structure
To be Published
|
|
2ARD
 
 | | The structure of tryptophan 7-halogenase (PrnA) suggests a mechanism for regioselective chlorination | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, tryptophan halogenase PrnA | | Authors: | Dong, C, Flecks, S, Unversucht, S, Haupt, C, Van Pee, K.H, Naismith, J.H, Scottish Structural Proteomics Facility (SSPF) | | Deposit date: | 2005-08-19 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Tryptophan 7-halogenase (PrnA) structure suggests a mechanism for regioselective chlorination.
Science, 309, 2005
|
|
6L1H
 
 | |
2APG
 
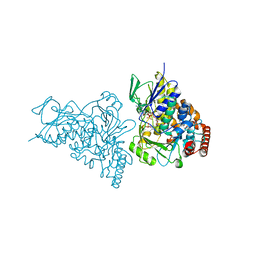 | | The structure of tryptophan 7-halogenase (PrnA)suggests a mechanism for regioselective chlorination | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Dong, C, Flecks, S, Unversucht, S, Haupt, C, Van Pee, K.H, Naismith, J.H, Scottish Structural Proteomics Facility (SSPF) | | Deposit date: | 2005-08-16 | | Release date: | 2005-10-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tryptophan 7-halogenase (PrnA) structure suggests a mechanism for regioselective chlorination.
Science, 309, 2005
|
|
2IYC
 
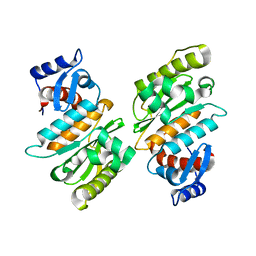 | | SENP1 native structure | | Descriptor: | SENTRIN-SPECIFIC PROTEASE 1 | | Authors: | Dong, C, Naismith, J.H. | | Deposit date: | 2006-07-14 | | Release date: | 2006-08-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Senp1 Native Structure
To be Published
|
|
2CKG
 
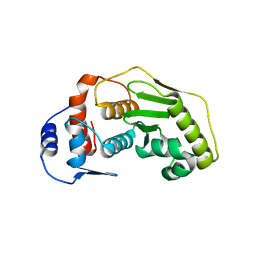 | |
6VCS
 
 | | SRA domain of UHRF1 in complex with DNA | | Descriptor: | DNA (5'-D(*GP*CP*CP*TP*GP*TP*AP*CP*AP*GP*GP*C)-3'), E3 ubiquitin-protein ligase UHRF1, UNK-UNK-UNK-UNK, ... | | Authors: | Dong, C, Tempel, W, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-12-22 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | SRA domain of UHRF1 in complex with DNA
To Be Published
|
|
5WP3
 
 | | Crystal Structure of EED in complex with EB22 | | Descriptor: | EB22, Polycomb protein EED, UNKNOWN ATOM OR ION | | Authors: | Dong, C, Tempel, W, Zhu, L, Moody, J.D, Baker, D, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-08-03 | | Release date: | 2017-09-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | First critical repressive H3K27me3 marks in embryonic stem cells identified using designed protein inhibitor.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6V8W
 
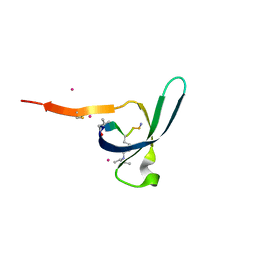 | | CDYL2 chromodomain in complex with a synthetic peptide | | Descriptor: | Chromodomain Y-like protein 2, IVA-PHE-ALA-PHE-5T3-SER-NH2, UNKNOWN ATOM OR ION | | Authors: | Dong, C, Tempel, W, James, L.I, Lamb, K.N, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Min, J, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-12-12 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
6CCT
 
 | | Fragment of GID4 in complex with a short peptide | | Descriptor: | Glucose-induced degradation protein 4 homolog, Tetrapeptide | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-07 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis of GID4-mediated recognition of degrons for the Pro/N-end rule pathway.
Nat. Chem. Biol., 14, 2018
|
|
5E1M
 
 | | Crystal structure of NTMT1 in complex with PPKRIA peptide | | Descriptor: | GLYCEROL, N-terminal Xaa-Pro-Lys N-methyltransferase 1, RCC1, ... | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-09-29 | | Release date: | 2015-10-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for substrate recognition by the human N-terminal methyltransferase 1.
Genes Dev., 29, 2015
|
|
5E2A
 
 | | Crystal structure of NTMT1 in complex with N-terminally methylated SPKRIA peptide | | Descriptor: | GLYCEROL, N-terminal Xaa-Pro-Lys N-methyltransferase 1, RCC1, ... | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-09-30 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for substrate recognition by the human N-terminal methyltransferase 1.
Genes Dev., 29, 2015
|
|
5E1D
 
 | | NTMT1 in complex with YPKRIA peptide | | Descriptor: | GLYCEROL, N-terminal Xaa-Pro-Lys N-methyltransferase 1, RCC1, ... | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-09-29 | | Release date: | 2015-10-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural basis for substrate recognition by the human N-terminal methyltransferase 1.
Genes Dev., 29, 2015
|
|
5E1O
 
 | | Crystal structure of NTMT1 in complex with RPKRIA peptide | | Descriptor: | GLYCEROL, N-terminal Xaa-Pro-Lys N-methyltransferase 1, RCC1, ... | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-09-29 | | Release date: | 2015-10-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for substrate recognition by the human N-terminal methyltransferase 1.
Genes Dev., 29, 2015
|
|
5E1B
 
 | | Crystal structure of NRMT1 in complex with SPKRIA peptide | | Descriptor: | GLYCEROL, N-terminal Xaa-Pro-Lys N-methyltransferase 1, RCC1, ... | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-09-29 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for substrate recognition by the human N-terminal methyltransferase 1.
Genes Dev., 29, 2015
|
|
5E2B
 
 | | Crystal structure of NTMT1 in complex with N-terminally methylated PPKRIA peptide | | Descriptor: | GLYCEROL, N-terminal Xaa-Pro-Lys N-methyltransferase 1, RCC1, ... | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-09-30 | | Release date: | 2015-10-28 | | Last modified: | 2015-12-02 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for substrate recognition by the human N-terminal methyltransferase 1.
Genes Dev., 29, 2015
|
|
2IXK
 
 | |
2IXJ
 
 | | RmlC P aeruginosa native | | Descriptor: | DTDP-4-DEHYDRORHAMNOSE 3,5-EPIMERASE, S,R MESO-TARTARIC ACID | | Authors: | Dong, C, Naismith, J.H. | | Deposit date: | 2006-07-08 | | Release date: | 2006-07-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Rmlc, a C3' and C5' Carbohydrate Epimerase, Appears to Operate Via an Intermediate with an Unusual Twist Boat Conformation.
J.Mol.Biol., 365, 2007
|
|
2IXI
 
 | | RmlC P aeruginosa with dTDP-xylose | | Descriptor: | DTDP-4-DEHYDRORHAMNOSE 3,5-EPIMERASE, S,R MESO-TARTARIC ACID, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Dong, C, Naismith, J.H. | | Deposit date: | 2006-07-08 | | Release date: | 2006-07-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rmlc, a C3' and C5' Carbohydrate Epimerase, Appears to Operate Via an Intermediate with an Unusual Twist Boat Conformation.
J.Mol.Biol., 365, 2007
|
|
6CDG
 
 | | GID4 fragment in complex with a peptide | | Descriptor: | Glucose-induced degradation protein 4 homolog, Hexapeptide PGLWKS, UNKNOWN ATOM OR ION | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-08 | | Release date: | 2018-03-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular basis of GID4-mediated recognition of degrons for the Pro/N-end rule pathway.
Nat. Chem. Biol., 14, 2018
|
|
6CD8
 
 | | Complex of GID4 fragment with short peptide | | Descriptor: | Glucose-induced degradation protein 4 homolog, Tetrapeptide PSRV, UNKNOWN ATOM OR ION | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-08 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular basis of GID4-mediated recognition of degrons for the Pro/N-end rule pathway.
Nat. Chem. Biol., 14, 2018
|
|
6CDC
 
 | | GID4 in complex with a tetrapeptide | | Descriptor: | Glucose-induced degradation protein 4 homolog, Tetrapeptide PGLW, UNKNOWN ATOM OR ION | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-08 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Molecular basis of GID4-mediated recognition of degrons for the Pro/N-end rule pathway.
Nat. Chem. Biol., 14, 2018
|
|
2IXH
 
 | | RmlC P aeruginosa with dTDP-rhamnose | | Descriptor: | 2'-DEOXY-THYMIDINE-BETA-L-RHAMNOSE, DTDP-4-DEHYDRORHAMNOSE 3,5-EPIMERASE | | Authors: | Dong, C, Naismith, J.H. | | Deposit date: | 2006-07-08 | | Release date: | 2006-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | RmlC, a C3' and C5' carbohydrate epimerase, appears to operate via an intermediate with an unusual twist boat conformation.
J. Mol. Biol., 365, 2007
|
|
6CCR
 
 | | Selenomethionyl derivative of a GID4 fragment | | Descriptor: | Glucose-induced degradation protein 4 homolog, UNKNOWN ATOM OR ION | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-07 | | Release date: | 2018-04-04 | | Last modified: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular basis of GID4-mediated recognition of degrons for the Pro/N-end rule pathway.
Nat. Chem. Biol., 14, 2018
|
|
