3MIW
 
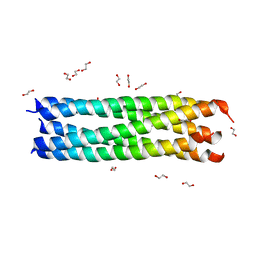 | | Crystal Structure of Rotavirus NSP4 | | Descriptor: | 1,2-ETHANEDIOL, Non-structural glycoprotein 4 | | Authors: | Chacko, A.R, Read, R.J, Dodson, E.J, Rao, D.C, Suguna, K. | | Deposit date: | 2010-04-12 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A new pentameric structure of rotavirus NSP4 revealed by molecular replacement.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2AAA
 
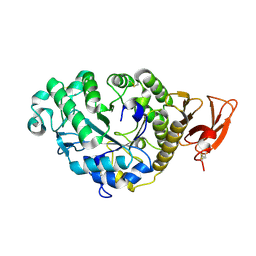 | | CALCIUM BINDING IN ALPHA-AMYLASES: AN X-RAY DIFFRACTION STUDY AT 2.1 ANGSTROMS RESOLUTION OF TWO ENZYMES FROM ASPERGILLUS | | Descriptor: | ALPHA-AMYLASE, CALCIUM ION | | Authors: | Brady, L, Brzozowski, A.M, Derewenda, Z, Dodson, E.J, Dodson, G.G. | | Deposit date: | 1991-02-27 | | Release date: | 1993-07-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Calcium binding in alpha-amylases: an X-ray diffraction study at 2.1-A resolution of two enzymes from Aspergillus.
Biochemistry, 29, 1990
|
|
1PMB
 
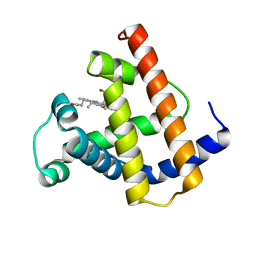 | | THE DETERMINATION OF THE CRYSTAL STRUCTURE OF RECOMBINANT PIG MYOGLOBIN BY MOLECULAR REPLACEMENT AND ITS REFINEMENT | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Smerdon, S.J, Oldfield, T.J, Dodson, E.J, Dodson, G.G, Hubbard, R.E, Wilkinson, A.J. | | Deposit date: | 1989-11-27 | | Release date: | 1990-01-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Determination of the crystal structure of recombinant pig myoglobin by molecular replacement and its refinement.
Acta Crystallogr.,Sect.B, 46, 1990
|
|
2BO4
 
 | | Dissection of mannosylglycerate synthase: an archetypal mannosyltransferase | | Descriptor: | CITRATE ANION, MANNOSYLGLYCERATE SYNTHASE | | Authors: | Flint, J, Taylor, E, Yang, M, Bolam, D.N, Tailford, L.E, Martinez-Fleites, C, Dodson, E.J, Davis, B.G, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2005-04-07 | | Release date: | 2005-06-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Dissection and High-Throughput Screening of Mannosylglyceerate Synthase
Nat.Struct.Mol.Biol., 12, 2005
|
|
2BSX
 
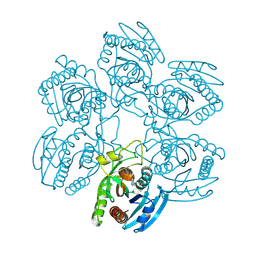 | | Crystal structure of the Plasmodium falciparum purine nucleoside phosphorylase complexed with inosine | | Descriptor: | INOSINE, PURINE NUCLEOSIDE PHOSPHORYLASE | | Authors: | Schnick, C, Brzozowski, A.M, Dodson, E.J, Murshudov, G.N, Brannigan, J.A, Wilkinson, A.J. | | Deposit date: | 2005-05-24 | | Release date: | 2005-08-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of Plasmodium Falciparum Purine Nucleoside Phosphorylase Complexed with Sulfate and its Natural Substrate Inosine
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2BO6
 
 | | DISSECTION OF MANNOSYLGLYCERATE SYNTHASE: AN ARCHETYPAL MANNOSYLTRANSFERASE | | Descriptor: | (2R)-2,3-DIHYDROXYPROPANOIC ACID, MANGANESE (II) ION, MANNOSYLGLYCERATE SYNTHASE | | Authors: | Flint, J, Taylor, E, Yang, M, Bolam, D.N, Tailford, L.E, Martinez-Fleites, C, Dodson, E.J, Davis, B.G, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2005-04-08 | | Release date: | 2005-06-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural Dissection and High-Throughput Screening of Mannosylglyceerate Synthase
Nat.Struct.Mol.Biol., 12, 2005
|
|
6ZM3
 
 | | The structure of an E2 ubiquitin-conjugating complex (UBC2-UEV1) essential for Leishmania amastigote differentiation | | Descriptor: | Putative ubiquitin-conjugating enzyme e2, Ubiquitin-conjugating enzyme-like protein | | Authors: | Burge, R.J, Dodson, E.J, Wilkinson, A.J, Mottram, J.C. | | Deposit date: | 2020-07-01 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Leishmania differentiation requires ubiquitin conjugation mediated by a UBC2-UEV1 E2 complex.
Plos Pathog., 16, 2020
|
|
2BO8
 
 | | DISSECTION OF MANNOSYLGLYCERATE SYNTHASE: AN ARCHETYPAL MANNOSYLTRANSFERASE | | Descriptor: | CHLORIDE ION, GUANOSINE 5'-(TRIHYDROGEN DIPHOSPHATE), P'-D-MANNOPYRANOSYL ESTER, ... | | Authors: | Flint, J, Taylor, E, Yang, M, Bolam, D.N, Tailford, L.E, Martinez-Fleites, C, Dodson, E.J, Davis, B.G, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2005-04-08 | | Release date: | 2005-06-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural dissection and high-throughput screening of mannosylglycerate synthase.
Nat. Struct. Mol. Biol., 12, 2005
|
|
2BO7
 
 | | DISSECTION OF MANNOSYLGLYCERATE SYNTHASE: AN ARCHETYPAL MANNOSYLTRANSFERASE | | Descriptor: | COBALT (II) ION, GUANOSINE-5'-DIPHOSPHATE, MANNOSYLGLYCERATE SYNTHASE | | Authors: | Flint, J, Taylor, E, Yang, M, Bolam, D.N, Tailford, L.E, Martinez-Fleites, C, Dodson, E.J, Davis, B.G, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2005-04-08 | | Release date: | 2005-06-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural dissection and high-throughput screening of mannosylglycerate synthase.
Nat. Struct. Mol. Biol., 12, 2005
|
|
1SAR
 
 | |
1RGP
 
 | | GTPASE-ACTIVATION DOMAIN FROM RHOGAP | | Descriptor: | RHOGAP | | Authors: | Barrett, T, Xiao, B, Dodson, E.J, Dodson, G, Ludbrook, S.B, Nurmahomed, K, Gamblin, S.J, Musacchio, A, Smerdon, S.J, Eccleston, J.F. | | Deposit date: | 1996-12-05 | | Release date: | 1997-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of the GTPase-activating domain from p50rhoGAP.
Nature, 385, 1997
|
|
1GYH
 
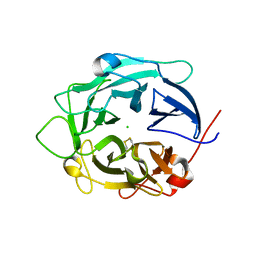 | | Structure of D158A Cellvibrio cellulosa alpha-L-arabinanase mutant | | Descriptor: | ARABINAN ENDO-1,5-ALPHA-L-ARABINOSIDASE A, CHLORIDE ION | | Authors: | Nurizzo, D, Turkenburg, J.P, Charnock, S.J, Roberts, S.M, Dodson, E.J, McKie, V.A, Taylor, E.J, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2002-04-23 | | Release date: | 2002-08-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Cellovibrio Cellulosa Alpha-L-Arabinanase 43A Has a Novel Five-Blade Beta-Propeller Fold
Nat.Struct.Biol., 9, 2002
|
|
1GYD
 
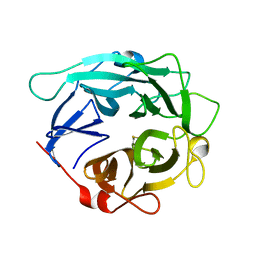 | | Structure of Cellvibrio cellulosa alpha-L-arabinanase | | Descriptor: | ARABINAN ENDO-1,5-ALPHA-L-ARABINOSIDASE A | | Authors: | Nurizzo, D, Turkenburg, J.P, Charnock, S.J, Roberts, S.M, Dodson, E.J, McKie, V.A, Taylor, E.J, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2002-04-23 | | Release date: | 2002-08-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Cellvibrio japonicus alpha-L-arabinanase 43A has a novel five-blade beta-propeller fold.
Nat. Struct. Biol., 9, 2002
|
|
1GYE
 
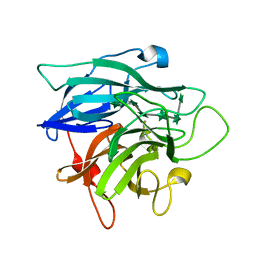 | | Structure of Cellvibrio cellulosa alpha-L-arabinanase complexed with Arabinohexaose | | Descriptor: | ARABINAN ENDO-1,5-ALPHA-L-ARABINOSIDASE A, CHLORIDE ION, alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose | | Authors: | Nurizzo, D, Turkenburg, J.P, Charnock, S.J, Roberts, S.M, Dodson, E.J, McKie, V.A, Taylor, E.J, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2002-04-23 | | Release date: | 2002-08-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Cellvibrio japonicus alpha-L-arabinanase 43A has a novel five-blade beta-propeller fold.
Nat. Struct. Biol., 9, 2002
|
|
7SMH
 
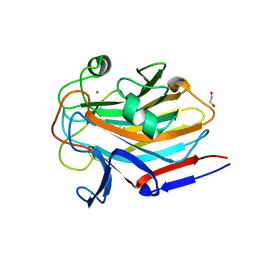 | | Structure of SASG A-domain (residues 163-419) from Staphylococcus aureus | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Surface protein G | | Authors: | Atkin, K.E, Whelan, F, Brentnall, A.S, Dodson, E.J, Turkenburg, J.P, Potts, J.R. | | Deposit date: | 2021-10-25 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Staphylococcal Periscope proteins Aap, SasG, and Pls project noncanonical legume-like lectin adhesin domains from the bacterial surface.
J.Biol.Chem., 299, 2023
|
|
1H02
 
 | | Human Insulin-like growth factor; SRS Daresbury data | | Descriptor: | INSULIN-LIKE GROWTH FACTOR I, N-DODECYL-N,N-DIMETHYL-3-AMMONIO-1-PROPANESULFONATE | | Authors: | Brzozowski, A.M, Dodson, E.J, Dodson, G.G, Murshudov, G, Verma, C, Turkenburg, J.P, De Bree, F.M, Dauter, Z. | | Deposit date: | 2002-06-11 | | Release date: | 2002-07-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Origins of the Functional Divergence of Human Insulin-Like Growth Factor-I and Insulin
Biochemistry, 41, 2002
|
|
1GZZ
 
 | | Human Insulin-like growth factor; Hamburg data | | Descriptor: | INSULIN-LIKE GROWTH FACTOR I, N-DODECYL-N,N-DIMETHYL-3-AMMONIO-1-PROPANESULFONATE | | Authors: | Brzozowski, A.M, Dodson, E.J, Dodson, G.G, Murshudov, G, Verma, C, Turkenburg, J.P, De Bree, F.M, Dauter, Z. | | Deposit date: | 2002-06-10 | | Release date: | 2002-07-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Origins of the Functional Divergence of Human Insulin-Like Growth Factor-I and Insulin
Biochemistry, 41, 2002
|
|
1GZR
 
 | | Human Insulin-like growth factor; ESRF data | | Descriptor: | INSULIN-LIKE GROWTH FACTOR I, N-DODECYL-N,N-DIMETHYL-3-AMMONIO-1-PROPANESULFONATE | | Authors: | Brzozowski, A.M, Dodson, E.J, Dodson, G.G, Murshudov, G, Verma, C, Turkenburg, J.P, De Bree, F.M, Dauter, Z. | | Deposit date: | 2002-05-28 | | Release date: | 2002-07-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Origins of the Functional Divergence of Human Insulin-Like Growth Factor-I and Insulin
Biochemistry, 41, 2002
|
|
1GZY
 
 | | Human Insulin-like growth factor; In-house data | | Descriptor: | INSULIN-LIKE GROWTH FACTOR I, N-DODECYL-N,N-DIMETHYL-3-AMMONIO-1-PROPANESULFONATE | | Authors: | Brzozowski, A.M, Dodson, E.J, Dodson, G.G, Murshudov, G, Verma, C, Turkenburg, J.P, De Bree, F.M, Dauter, Z. | | Deposit date: | 2002-06-10 | | Release date: | 2002-07-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structural Origins of the Functional Divergence of Human Insulin-Like Growth Factor-I and Insulin
Biochemistry, 41, 2002
|
|
1K0R
 
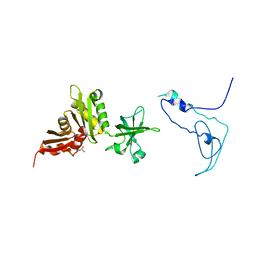 | | Crystal Structure of Mycobacterium tuberculosis NusA | | Descriptor: | NusA, SULFATE ION | | Authors: | Gopal, B, Haire, L.F, Gamblin, S.J, Dodson, E.J, Lane, A.N, Papavinasasundaram, K.G, Colston, M.J, Dodson, G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2001-09-20 | | Release date: | 2001-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the transcription elongation/anti-termination factor NusA from Mycobacterium tuberculosis at 1.7 A resolution.
J.Mol.Biol., 314, 2001
|
|
8AY0
 
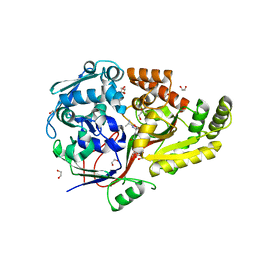 | | Crystal Structure of the peptide binding protein DppE from Bacillus subtilis in complex with murein tripeptide | | Descriptor: | 1,2-ETHANEDIOL, Dipeptide-binding protein DppE, L-ALA-GAMMA-D-GLU-MESO-DIAMINOPIMELIC ACID, ... | | Authors: | Hughes, A.M, Dodson, E.J, Wilkinson, A.J. | | Deposit date: | 2022-09-01 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Peptide transport in Bacillus subtilis - structure and specificity in the extracellular solute binding proteins OppA and DppE.
Microbiology (Reading, Engl.), 168, 2022
|
|
8ARN
 
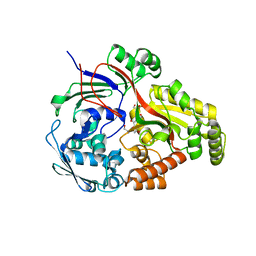 | | Crystal structure of the peptide binding protein, OppA, from Bacillus subtilis in complex with an endogenous tetrapeptide | | Descriptor: | Endogenous tetrapeptide (SER-ASN-SER-SER), Oligopeptide-binding protein OppA | | Authors: | Hughes, A, Dodson, E.J, Wilkinson, A.J. | | Deposit date: | 2022-08-17 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Peptide transport in Bacillus subtilis - structure and specificity in the extracellular solute binding proteins OppA and DppE.
Microbiology (Reading, Engl.), 168, 2022
|
|
8ARE
 
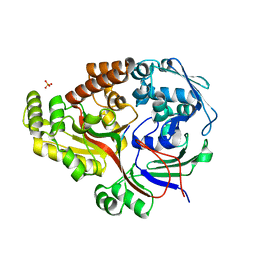 | | Crystal structure of the peptide binding protein, OppA, from Bacillus subtilis in complex with a PhrE-derived pentapeptide | | Descriptor: | Oligopeptide-binding protein OppA, Phosphatase RapE inhibitor, SULFATE ION | | Authors: | Hughes, A, Dodson, E.J, Wilkinson, A.J. | | Deposit date: | 2022-08-16 | | Release date: | 2023-02-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Peptide transport in Bacillus subtilis - structure and specificity in the extracellular solute binding proteins OppA and DppE.
Microbiology (Reading, Engl.), 168, 2022
|
|
8AZB
 
 | |
8BAW
 
 | | X-ray structure of the CeuE Homologue from Geobacillus stearothermophilus - 5-LICAM siderophore analogue complex. | | Descriptor: | FE (III) ION, N,N'-pentane-1,5-diylbis(2,3-dihydroxybenzamide), Siderophore ABC transporter substrate-binding protein | | Authors: | Blagova, E.V, Miller, A, Booth, R, Dodson, E.J, Duhme-Klair, A.K, Wilson, K.S. | | Deposit date: | 2022-10-12 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.471 Å) | | Cite: | Thermostable homologues of the periplasmic siderophore-binding protein CeuE from Geobacillus stearothermophilus and Parageobacillus thermoglucosidasius.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
