3KNQ
 
 | |
3KJG
 
 | | ADP-bound state of CooC1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CO dehydrogenase/acetyl-CoA synthase complex, accessory protein CooC | | Authors: | Jeoung, J.-H, Dobbek, H. | | Deposit date: | 2009-11-03 | | Release date: | 2010-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the ATP-Dependent Maturation Factor of Ni,Fe-Containing Carbon Monoxide Dehydrogenases
J.Mol.Biol., 396, 2010
|
|
2X9B
 
 | | The filamentous phages fd and IF1 use different infection mechanisms | | Descriptor: | ATTACHMENT PROTEIN G3P | | Authors: | Lorenz, S.H, Jakob, R.P, Weininger, U, Dobbek, H, Schmid, F.X. | | Deposit date: | 2010-03-15 | | Release date: | 2010-12-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | The Filamentous Phages Fd and If1 Use Different Mechanisms to Infect Escherichia Coli.
J.Mol.Biol., 405, 2011
|
|
4PJ0
 
 | | Structure of T.elongatus Photosystem II, rows of dimers crystal packing | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Hellmich, J, Bommer, M, Burkhardt, A, Ibrahim, M, Kern, J, Meents, A, Mueh, F, Dobbek, H, Zouni, A. | | Deposit date: | 2014-05-10 | | Release date: | 2014-10-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.437 Å) | | Cite: | Native-like Photosystem II Superstructure at 2.44 angstrom Resolution through Detergent Extraction from the Protein Crystal.
Structure, 22, 2014
|
|
3PVJ
 
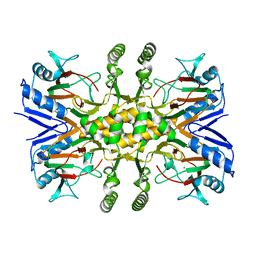 | | Crystal structure of the Fe(II)/alpha-ketoglutarate dependent taurine dioxygenase from Pseudomonas putida KT2440 | | Descriptor: | Alpha-ketoglutarate-dependent taurine dioxygenase, CHLORIDE ION | | Authors: | Knauer, S.H, Spiegelhauer, O, Schwarzinger, S, Haenzelmann, P, Dobbek, H. | | Deposit date: | 2010-12-07 | | Release date: | 2011-12-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Biochemical and structural characterization of the Fe(II)/alpha-ketoglutarate dependent taurine dioxygenase from Pseudomonas putida KT2440
To be Published
|
|
5AA5
 
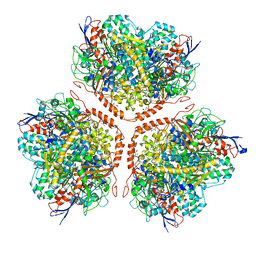 | | Actinobacterial-type NiFe-hydrogenase from Ralstonia eutropha H16 at 2.85 Angstrom resolution | | Descriptor: | IRON/SULFUR CLUSTER, MALONIC ACID, NIFE-HYDROGENASE LARGE SUBUNIT, ... | | Authors: | Schaefer, C, Bommer, M, Hennig, S, Jeoung, J.H, Dobbek, H, Lenz, O. | | Deposit date: | 2015-07-23 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Structure of an Actinobacterial-Type [Nife]-Hydrogenase Reveals Insight Into O2-Tolerant H2 Oxidation.
Structure, 24, 2016
|
|
4UTH
 
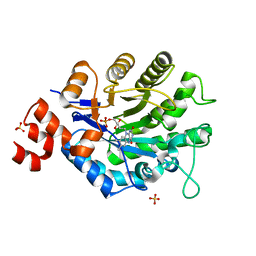 | | XenA - oxidized - Y183F variant | | Descriptor: | FLAVIN MONONUCLEOTIDE, NADH:flavin oxidoreductase, SULFATE ION | | Authors: | Werther, T, Dobbek, H. | | Deposit date: | 2014-07-21 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Redox-dependent substrate-cofactor interactions in the Michaelis-complex of a flavin-dependent oxidoreductase
Nat Commun, 8, 2017
|
|
6G2G
 
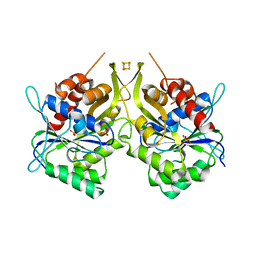 | | Fe-S assembly Cfd1 | | Descriptor: | Cytosolic Fe-S cluster assembly factor CFD1, IRON/SULFUR CLUSTER, SULFATE ION | | Authors: | Jeoung, J.H, Dobbek, H. | | Deposit date: | 2018-03-23 | | Release date: | 2019-01-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Function and crystal structure of the dimeric P-loop ATPase CFD1 coordinating an exposed [4Fe-4S] cluster for transfer to apoproteins.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1FFV
 
 | | CARBON MONOXIDE DEHYDROGENASE FROM HYDROGENOPHAGA PSEUDOFLAVA | | Descriptor: | (MOLYBDOPTERIN-CYTOSINE DINUCLEOTIDE-S,S)-DIOXO-AQUA-MOLYBDENUM(V), CUTL, MOLYBDOPROTEIN OF CARBON MONOXIDE DEHYDROGENASE, ... | | Authors: | Haenzelmann, P, Dobbek, H, Gremer, L, Huber, R, Meyer, O. | | Deposit date: | 2000-07-26 | | Release date: | 2000-09-15 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The effect of intracellular molybdenum in Hydrogenophaga pseudoflava on the crystallographic structure of the seleno-molybdo-iron-sulfur flavoenzyme carbon monoxide dehydrogenase.
J.Mol.Biol., 301, 2000
|
|
1FFU
 
 | | CARBON MONOXIDE DEHYDROGENASE FROM HYDROGENOPHAGA PSEUDOFLAVA WHICH LACKS THE MO-PYRANOPTERIN MOIETY OF THE MOLYBDENUM COFACTOR | | Descriptor: | CUTL, MOLYBDOPROTEIN OF CARBON MONOXIDE DEHYDROGENASE, CUTM, ... | | Authors: | Haenzelmann, P, Dobbek, H, Gremer, L, Huber, R, Meyer, O. | | Deposit date: | 2000-07-26 | | Release date: | 2000-09-15 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The effect of intracellular molybdenum in Hydrogenophaga pseudoflava on the crystallographic structure of the seleno-molybdo-iron-sulfur flavoenzyme carbon monoxide dehydrogenase.
J.Mol.Biol., 301, 2000
|
|
3I39
 
 | | NI,FE-CODH-320 MV+CN state | | Descriptor: | CYANIDE ION, Carbon monoxide dehydrogenase 2, FE (II) ION, ... | | Authors: | Jeoung, J.-H, Dobbek, H. | | Deposit date: | 2009-06-30 | | Release date: | 2009-08-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structural basis of cyanide inhibition of Ni, Fe-containing carbon monoxide dehydrogenase
J.Am.Chem.Soc., 131, 2009
|
|
7B9A
 
 | | CooS-V with Xe-soaked | | Descriptor: | 3,5-dioxa-7-thia-1-thionia-2$l^{2},4$l^{2},6$l^{3},8$l^{2}-tetraferrabicyclo[4.2.0]octane, BROMIDE ION, Carbon monoxide dehydrogenase, ... | | Authors: | Jeoung, J.H, Dobbek, H. | | Deposit date: | 2020-12-14 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Morphing [4Fe-3S-nO]-Cluster within a Carbon Monoxide Dehydrogenase Scaffold.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
3HRD
 
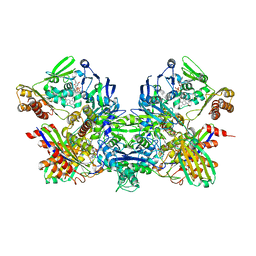 | | Crystal structure of nicotinate dehydrogenase | | Descriptor: | CALCIUM ION, DIOXOTHIOMOLYBDENUM(VI) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Wagener, N, Pierik, A.J, Hille, R, Dobbek, H. | | Deposit date: | 2009-06-09 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Mo-Se active site of nicotinate dehydrogenase
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3DGS
 
 | |
5KAF
 
 | | RT XFEL structure of Photosystem II in the dark state at 3.0 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Young, I.D, Ibrahim, M, Chatterjee, R, Gul, S, Koroidov, S, Brewster, A.S, Tran, R, Alonso-Mori, R, Fuller, F, Kroll, T, Michels-Clark, T, Laksmono, H, Sierra, R.G, Stan, C.A, Saracini, C, Bean, M.A, Seuffert, I, Sokaras, D, Weng, T.-C, Hunter, M.S, Aquila, A, Koglin, J.E, Robinson, J, Liang, M, Boutet, S, Lyubimov, A.Y, Uervirojnangkoorn, M, Moriarty, N.W, Liebschner, D, Afonine, P.V, Waterman, D.G, Evans, G, Dobbek, H, Weis, W.I, Brunger, A.T, Zwart, P.H, Adams, P.D, Zouni, A, Messinger, J, Bergmann, U, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2016-06-01 | | Release date: | 2016-11-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.00001 Å) | | Cite: | Structure of photosystem II and substrate binding at room temperature.
Nature, 540, 2016
|
|
5KAI
 
 | | NH3-bound RT XFEL structure of Photosystem II 500 ms after the 2nd illumination (2F) at 2.8 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Young, I.D, Ibrahim, M, Chatterjee, R, Gul, S, Koroidov, S, Brewster, A.S, Tran, R, Alonso-Mori, R, Fuller, F, Kroll, T, Michels-Clark, T, Laksmono, H, Sierra, R.G, Stan, C.A, Saracini, C, Bean, M.A, Seuffert, I, Sokaras, D, Weng, T.-C, Hunter, M.S, Aquila, A, Koglin, J.E, Robinson, J, Liang, M, Boutet, S, Lyubimov, A.Y, Uervirojnangkoorn, M, Moriarty, N.W, Liebschner, D, Afonine, P.V, Waterman, D.G, Evans, G, Dobbek, H, Weis, W.I, Brunger, A.T, Zwart, P.H, Adams, P.D, Zouni, A, Messinger, J, Bergmann, U, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2016-06-01 | | Release date: | 2016-11-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.80000925 Å) | | Cite: | Structure of photosystem II and substrate binding at room temperature.
Nature, 540, 2016
|
|
8CJB
 
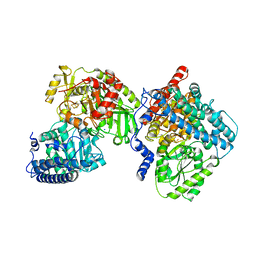 | | A268M variant of the CODH/ACS complex of C. hydrogenoformans | | Descriptor: | ACETATE ION, CO-methylating acetyl-CoA synthase, Carbon monoxide dehydrogenase, ... | | Authors: | Ruickoldt, J, Jeoung, J, Lennartz, F, Dobbek, H. | | Deposit date: | 2023-02-13 | | Release date: | 2024-02-21 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Coupling CO 2 Reduction and Acetyl-CoA Formation: The Role of a CO Capturing Tunnel in Enzymatic Catalysis.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8CJA
 
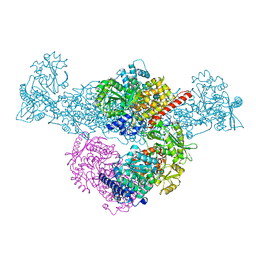 | | A225L/F231A variant of the CODH/ACS complex of C. hydrogenoformans | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CO-methylating acetyl-CoA synthase, ... | | Authors: | Ruickoldt, J, Jeoung, J, Lennartz, F, Dobbek, H. | | Deposit date: | 2023-02-13 | | Release date: | 2024-02-21 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Coupling CO 2 Reduction and Acetyl-CoA Formation: The Role of a CO Capturing Tunnel in Enzymatic Catalysis.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8CMW
 
 | | A225L variant of the CODH/ACS complex of C. hydrogenoformans | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CO-methylating acetyl-CoA synthase, ... | | Authors: | Ruickoldt, J, Jeoung, J, Lennartz, F, Dobbek, H. | | Deposit date: | 2023-02-21 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Coupling CO 2 Reduction and Acetyl-CoA Formation: The Role of a CO Capturing Tunnel in Enzymatic Catalysis.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
3DTM
 
 | |
8CJC
 
 | | F515A variant of the CODH/ACS complex of C. hydrogenoformans | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CO-methylating acetyl-CoA synthase, ... | | Authors: | Ruickoldt, J, Jeoung, J, Lennartz, F, Dobbek, H. | | Deposit date: | 2023-02-13 | | Release date: | 2024-02-21 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Coupling CO 2 Reduction and Acetyl-CoA Formation: The Role of a CO Capturing Tunnel in Enzymatic Catalysis.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8OMY
 
 | | NI,FE-CODH -600mV state : 35 min Dioxygen Exposure | | Descriptor: | Carbon monoxide dehydrogenase 2, FE (II) ION, FE (III) ION, ... | | Authors: | Basak, Y, Jeoung, J.-H, Dobbek, H. | | Deposit date: | 2023-03-31 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Stepwise O 2 -Induced Rearrangement and Disassembly of the [NiFe 4 (OH)( mu 3 -S) 4 ] Active Site Cluster of CO Dehydrogenase.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8ON2
 
 | | NI,FE-CODH -600mV state : 24 h Dioxygen Exposure | | Descriptor: | Carbon monoxide dehydrogenase 2, FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Basak, Y, Jeoung, J.-H, Dobbek, H. | | Deposit date: | 2023-03-31 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Stepwise O 2 -Induced Rearrangement and Disassembly of the [NiFe 4 (OH)( mu 3 -S) 4 ] Active Site Cluster of CO Dehydrogenase.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8OMX
 
 | | NI,FE-CODH -600mV state : 1 min Dioxygen Exposure | | Descriptor: | Carbon monoxide dehydrogenase 2, FE (II) ION, FE(4)-NI(1)-S(5) CLUSTER, ... | | Authors: | Basak, Y, Jeoung, J.-H, Dobbek, H. | | Deposit date: | 2023-03-31 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Stepwise O 2 -Induced Rearrangement and Disassembly of the [NiFe 4 (OH)( mu 3 -S) 4 ] Active Site Cluster of CO Dehydrogenase.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8ON1
 
 | | NI,FE-CODH -600mV state : 4 h Dioxygen Exposure | | Descriptor: | Carbon monoxide dehydrogenase 2, FE (II) ION, FE (III) ION, ... | | Authors: | Basak, Y, Jeoung, J.-H, Dobbek, H. | | Deposit date: | 2023-03-31 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Stepwise O 2 -Induced Rearrangement and Disassembly of the [NiFe 4 (OH)( mu 3 -S) 4 ] Active Site Cluster of CO Dehydrogenase.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
