5MAU
 
 | | Crystal structure of dimeric chlorite dismutase from Cyanothece sp. PCC7425 (pH 6.5) | | Descriptor: | Chlorite dismutase, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Puehringer, D, Schaffner, I, Mlynek, G, Obinger, C, Djinovic-Carugo, K. | | Deposit date: | 2016-11-04 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Molecular Mechanism of Enzymatic Chlorite Detoxification: Insights from Structural and Kinetic Studies.
ACS Catal, 7, 2017
|
|
5NKU
 
 | | Joint neutron/X-ray structure of dimeric chlorite dismutase from Cyanothece sp. PCC7425 | | Descriptor: | CHLORIDE ION, Chlorite Dismutase, GLYCEROL, ... | | Authors: | Puehringer, D, Schaffner, I, Mlynek, G, Obinger, C, Djinovic-Carugo, K. | | Deposit date: | 2017-04-03 | | Release date: | 2018-02-28 | | Last modified: | 2024-05-01 | | Method: | NEUTRON DIFFRACTION (2 Å), X-RAY DIFFRACTION | | Cite: | Molecular Mechanism of Enzymatic Chlorite Detoxification: Insights from Structural and Kinetic Studies.
ACS Catal, 7, 2017
|
|
1TJT
 
 | |
1WKU
 
 | |
5JWG
 
 | | Crystal structure of Porphyromonas endodontalis DPP11 in complex with dipeptide Arg-Asp | | Descriptor: | 1,2-ETHANEDIOL, ARGININE, ASPARTIC ACID, ... | | Authors: | Bezerra, G.A, Fedosyuk, S, Ohara-Nemoto, Y, Nemoto, T.K, Djinovic-Carugo, K. | | Deposit date: | 2016-05-12 | | Release date: | 2017-06-14 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bacterial protease uses distinct thermodynamic signatures for substrate recognition.
Sci Rep, 7, 2017
|
|
5JXF
 
 | | Crystal structure of Flavobacterium psychrophilum DPP11 in complex with dipeptide Arg-Asp | | Descriptor: | ARGININE, ASPARTIC ACID, Asp/Glu-specific dipeptidyl-peptidase, ... | | Authors: | Bezerra, G.A, Fedosyuk, S, Ohara-Nemoto, Y, Nemoto, T.K, Djinovic-Carugo, K. | | Deposit date: | 2016-05-13 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Bacterial protease uses distinct thermodynamic signatures for substrate recognition.
Sci Rep, 7, 2017
|
|
5JWF
 
 | | Crystal structure of Porphyromonas gingivalis DPP11 | | Descriptor: | Asp/Glu-specific dipeptidyl-peptidase, GLYCEROL, S-1,2-PROPANEDIOL | | Authors: | Bezerra, G.A, Fedosyuk, S, Ohara-Nemoto, Y, Nemoto, T.K, Djinovic-Carugo, K. | | Deposit date: | 2016-05-12 | | Release date: | 2017-06-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Bacterial protease uses distinct thermodynamic signatures for substrate recognition.
Sci Rep, 7, 2017
|
|
5K33
 
 | | Crystal structure of extracellular domain of HER2 in complex with Fcab STAB19 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Ig gamma-1 chain C region, ... | | Authors: | Humm, A, Lobner, E, Goritzer, K, Mlynek, G, Obinger, C, Djinovic-Carugo, K. | | Deposit date: | 2016-05-19 | | Release date: | 2017-04-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Fcab-HER2 Interaction: a Menage a Trois. Lessons from X-Ray and Solution Studies.
Structure, 25, 2017
|
|
5JWI
 
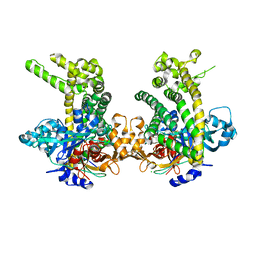 | | Crystal structure of Porphyromonas endodontalis DPP11 in complex with dipeptide Arg-Glu | | Descriptor: | ARGININE, Asp/Glu-specific dipeptidyl-peptidase, CHLORIDE ION, ... | | Authors: | Bezerra, G.A, Fedosyuk, S, Ohara-Nemoto, Y, Nemoto, T.K, Djinovic-Carugo, K. | | Deposit date: | 2016-05-12 | | Release date: | 2017-06-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Bacterial protease uses distinct thermodynamic signatures for substrate recognition.
Sci Rep, 7, 2017
|
|
5KWG
 
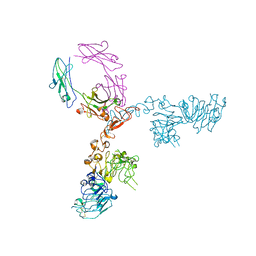 | | Crystal structure of extracellular domain of HER2 in complex with Fcab H10-03-6 | | Descriptor: | Ig gamma-1 chain C region, Receptor tyrosine-protein kinase erbB-2 | | Authors: | Humm, A, Lobner, E, Goritzer, K, Mlynek, G, Obinger, C, Djinovic-Carugo, K. | | Deposit date: | 2016-07-18 | | Release date: | 2017-04-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Fcab-HER2 Interaction: a Menage a Trois. Lessons from X-Ray and Solution Studies.
Structure, 25, 2017
|
|
5K65
 
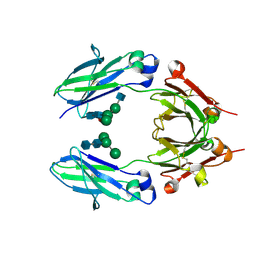 | | Crystal structure of VEGF binding IgG1-Fc (Fcab CT6) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Ig gamma-1 chain C region, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Humm, A, Lobner, E, Kitzmuller, M, Mlynek, G, Obinger, C, Djinovic-Carugo, K. | | Deposit date: | 2016-05-24 | | Release date: | 2017-09-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Two-faced Fcab prevents polymerization with VEGF and reveals thermodynamics and the 2.15 angstrom crystal structure of the complex.
MAbs, 9, 2017
|
|
5K8Z
 
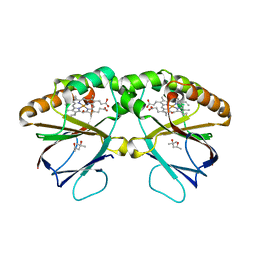 | | Crystal structure of dimeric chlorite dismutase from Cyanothece sp. PCC7425 (pH 8.5) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, Chlorite dismutase, ... | | Authors: | Puehringer, D, Schaffner, I, Mlynek, G, Obinger, C, Djinovic-Carugo, K. | | Deposit date: | 2016-05-31 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Molecular Mechanism of Enzymatic Chlorite Detoxification: Insights from Structural and Kinetic Studies.
ACS Catal, 7, 2017
|
|
6RWV
 
 | | Structure of apo-LmCpfC | | Descriptor: | Ferrochelatase, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Hofbauer, S, Helm, J, Djinovic-Carugo, K, Furtmueller, P.G. | | Deposit date: | 2019-06-06 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6386379 Å) | | Cite: | Crystal structures and calorimetry reveal catalytically relevant binding mode of coproporphyrin and coproheme in coproporphyrin ferrochelatase.
Febs J., 287, 2020
|
|
5JY0
 
 | | Crystal structure of Porphyromonas endodontalis DPP11 in complex with substrate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Asp/Glu-specific dipeptidyl-peptidase, LEU-ASP-VAL | | Authors: | Bezerra, G.A, Cornaciu, I, Hoffmann, G, Djinovic-Carugo, K, Marquez, J.A. | | Deposit date: | 2016-05-13 | | Release date: | 2017-06-14 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (2.599 Å) | | Cite: | Bacterial protease uses distinct thermodynamic signatures for substrate recognition.
Sci Rep, 7, 2017
|
|
5JXP
 
 | | Crystal structure of Porphyromonas endodontalis DPP11 in alternate conformation | | Descriptor: | Asp/Glu-specific dipeptidyl-peptidase, CALCIUM ION, CHLORIDE ION | | Authors: | Bezerra, G.A, Cornaciu, I, Hoffmann, G, Djinovic-Carugo, K, Marquez, J.A. | | Deposit date: | 2016-05-13 | | Release date: | 2017-06-14 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Bacterial protease uses distinct thermodynamic signatures for substrate recognition.
Sci Rep, 7, 2017
|
|
5K90
 
 | | Crystal structure of dimeric chlorite dismutase from Cyanothece sp. PCC7425 in complex with isothiocyanate | | Descriptor: | Chlorite dismutase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Puehringer, D, Schaffner, I, Mlynek, G, Obinger, C, Djinovic-Carugo, K. | | Deposit date: | 2016-05-31 | | Release date: | 2017-06-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Molecular Mechanism of Enzymatic Chlorite Detoxification: Insights from Structural and Kinetic Studies.
ACS Catal, 7, 2017
|
|
5K64
 
 | | Crystal structure of VEGF binding IgG1-Fc (Fcab 448) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Humm, A, Lobner, E, Kitzmuller, M, Mlynek, G, Obinger, C, Djinovic-Carugo, K. | | Deposit date: | 2016-05-24 | | Release date: | 2017-09-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Two-faced Fcab prevents polymerization with VEGF and reveals thermodynamics and the 2.15 angstrom crystal structure of the complex.
MAbs, 9, 2017
|
|
5K91
 
 | | Crystal structure of dimeric chlorite dismutase from Cyanothece sp. PCC7425 in complex with fluoride | | Descriptor: | Chlorite dismutase, FLUORIDE ION, GLYCEROL, ... | | Authors: | Puehringer, D, Schaffner, I, Mlynek, G, Obinger, C, Djinovic-Carugo, K. | | Deposit date: | 2016-05-31 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Molecular Mechanism of Enzymatic Chlorite Detoxification: Insights from Structural and Kinetic Studies.
ACS Catal, 7, 2017
|
|
6T69
 
 | | Crystal structure of Toxoplasma gondii Morn1(V shape) | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Membrane occupation and recognition nexus protein MORN1, ... | | Authors: | Grishkovskaya, I, Kostan, J, Sajko, S, Morriswood, B, Djinovic-Carugo, K. | | Deposit date: | 2019-10-18 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of three MORN repeat proteins and a re-evaluation of the proposed lipid-binding properties of MORN repeats.
Plos One, 15, 2020
|
|
6T4D
 
 | | Crystal structure of Plasmodium falciparum Morn1 | | Descriptor: | Morn1, ZINC ION | | Authors: | Grishkovskaya, I, Kostan, J, Sajko, S, Morriswood, B, Djinovic-Carugo, K. | | Deposit date: | 2019-10-13 | | Release date: | 2020-11-18 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structures of three MORN repeat proteins and a re-evaluation of the proposed lipid-binding properties of MORN repeats.
Plos One, 15, 2020
|
|
6T6Q
 
 | | Crystal structure of Toxoplasma gondii Morn1 (extended conformation). | | Descriptor: | Membrane occupation and recognition nexus protein MORN1 | | Authors: | Grishkovskaya, I, Kostan, J, Sajko, S, Morriswood, B, Djinovic-Carugo, K. | | Deposit date: | 2019-10-18 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.902 Å) | | Cite: | Structures of three MORN repeat proteins and a re-evaluation of the proposed lipid-binding properties of MORN repeats.
Plos One, 15, 2020
|
|
5JIH
 
 | | Crystal structure of HER2 binding IgG1-Fc (Fcab STAB19) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ig gamma-1 chain C region | | Authors: | Humm, A, Lobner, E, Mlynek, G, Obinger, C, Djinovic-Carugo, K. | | Deposit date: | 2016-04-22 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.663 Å) | | Cite: | Fcab-HER2 Interaction: a Menage a Trois. Lessons from X-Ray and Solution Studies.
Structure, 25, 2017
|
|
6T68
 
 | | Crystal structure of Trypanosoma brucei Morn1 | | Descriptor: | CHLORIDE ION, MORN repeat-containing protein 1 | | Authors: | Grishkovskaya, I, Kostan, J, Sajko, S, Morriswood, B, Djinovic-Carugo, K. | | Deposit date: | 2019-10-17 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structures of three MORN repeat proteins and a re-evaluation of the proposed lipid-binding properties of MORN repeats.
Plos One, 15, 2020
|
|
5JXK
 
 | | Crystal structure of Porphyromonas endodontalis DPP11 | | Descriptor: | Asp/Glu-specific dipeptidyl-peptidase, CHLORIDE ION | | Authors: | Bezerra, G.A, Cornaciu, I, Hoffmann, G, Djinovic-Carugo, K, Marquez, J.A. | | Deposit date: | 2016-05-13 | | Release date: | 2017-06-14 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Bacterial protease uses distinct thermodynamic signatures for substrate recognition.
Sci Rep, 7, 2017
|
|
5JIK
 
 | | Crystal structure of HER2 binding IgG1-Fc (Fcab H10-03-6) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ig gamma-1 chain C region, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Humm, A, Lobner, E, Mlynek, G, Obinger, C, Djinovic-Carugo, K. | | Deposit date: | 2016-04-22 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Fcab-HER2 Interaction: a Menage a Trois. Lessons from X-Ray and Solution Studies.
Structure, 25, 2017
|
|
