2QE0
 
 | | Thioacylenzyme Intermediate of GAPN from S. Mutans, New Data Integration and Refinement. | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-dependent glyceraldehyde-3-phosphate dehydrogenase | | Authors: | Corbier, C, Didierjean, C, Bricogne, G, Branlant, G, D'Ambrosio, K, Vonrhein, C. | | Deposit date: | 2007-06-22 | | Release date: | 2007-12-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The First Crystal Structure of a Thioacylenzyme Intermediate in the ALDH Family: New Coenzyme Conformation and Relevance to Catalysis
Biochemistry, 45, 2006
|
|
4G10
 
 | | LigG from Sphingobium sp. SYK-6 is related to the glutathione transferase omega class | | Descriptor: | ACETATE ION, GLUTATHIONE, Glutathione S-transferase homolog, ... | | Authors: | Meux, E, Prosper, P, Masai, E, Mulliert Carlin, G, Dumarcay, S, Morel, M, Didierjean, C, Gelhaye, E, Favier, F. | | Deposit date: | 2012-07-10 | | Release date: | 2012-10-03 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Sphingobium sp. SYK-6 LigG involved in lignin degradation is structurally and biochemically related to the glutathione transferase omega class.
Febs Lett., 586, 2012
|
|
2X5J
 
 | | Crystal structure of the Apoform of the D-Erythrose-4-phosphate dehydrogenase from E. coli | | Descriptor: | D-ERYTHROSE-4-PHOSPHATE DEHYDROGENASE, PHOSPHATE ION | | Authors: | Moniot, S, Didierjean, C, Boschi-Muller, S, Branlant, G, Corbier, C. | | Deposit date: | 2010-02-09 | | Release date: | 2011-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Characterization of Erythrose-4- Phosphate Dehydrogenase from Escherichia Coli: Peculiar Features When Compared to Phosphorylating Gapdhs
To be Published
|
|
2XF8
 
 | | Structure of the D-Erythrose-4-Phosphate Dehydrogenase from E. coli in complex with a NAD cofactor analog (3-Chloroacetyl adenine pyridine dinucleotide) and sulfate anion | | Descriptor: | 3-(CHLOROACETYL) PYRIDINE ADENINE DINUCLEOTIDE, D-ERYTHROSE-4-PHOSPHATE DEHYDROGENASE, SULFATE ION | | Authors: | Moniot, S, Didierjean, C, Boschi-Muller, S, Branlant, G, Corbier, C. | | Deposit date: | 2010-05-20 | | Release date: | 2011-06-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural Characterization of Erythrose-4- Phosphate Dehydrogenase from Escherichia Coli: Peculiar Features When Compared to Phosphorylating Gapdhs
To be Published
|
|
2X5K
 
 | | Structure of an active site mutant of the D-Erythrose-4-Phosphate Dehydrogenase from E. coli | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, D-ERYTHROSE-4-PHOSPHATE DEHYDROGENASE, ... | | Authors: | Moniot, S, Didierjean, C, Boschi-Muller, S, Branlant, G, Corbier, C. | | Deposit date: | 2010-02-10 | | Release date: | 2011-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural Characterization of Erythrose-4- Phosphate Dehydrogenase from Escherichia Coli: Peculiar Features When Compared to Phosphorylating Gapdhs
To be Published
|
|
2P5R
 
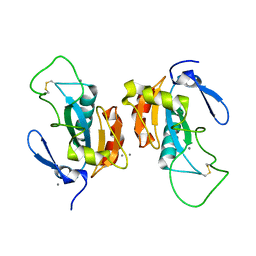 | | Crystal structure of the poplar glutathione peroxidase 5 in the oxidized form | | Descriptor: | CALCIUM ION, Glutathione peroxidase 5 | | Authors: | Koh, C.S, Didierjean, C, Navrot, N, Panjikar, S, Mulliert, G, Rouhier, N, Jacquot, J.-P, Aubry, A, Shawkataly, O, Corbier, C. | | Deposit date: | 2007-03-16 | | Release date: | 2007-07-24 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structures of a Poplar Thioredoxin Peroxidase that Exhibits the Structure of Glutathione Peroxidases: Insights into Redox-driven Conformational Changes.
J.Mol.Biol., 370, 2007
|
|
2P5Q
 
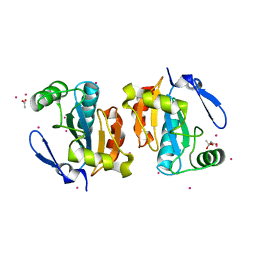 | | Crystal structure of the poplar glutathione peroxidase 5 in the reduced form | | Descriptor: | ACETATE ION, CADMIUM ION, Glutathione peroxidase 5 | | Authors: | Koh, C.S, Didierjean, C, Navrot, N, Panjikar, S, Mulliert, G, Rouhier, N, Jacquot, J.-P, Aubry, A, Shawkataly, O, Corbier, C. | | Deposit date: | 2007-03-16 | | Release date: | 2007-07-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of a Poplar Thioredoxin Peroxidase that Exhibits the Structure of Glutathione Peroxidases: Insights into Redox-driven Conformational Changes.
J.Mol.Biol., 370, 2007
|
|
8AX2
 
 | |
8AWZ
 
 | |
8AX0
 
 | |
8AX1
 
 | |
2MMA
 
 | | NMR-based docking model of GrxS14-BolA2 apo-heterodimer from Arabidopsis thaliana | | Descriptor: | BolA2, Monothiol glutaredoxin-S14, chloroplastic | | Authors: | Roret, T, Tsan, P, Couturier, J, Rouhier, N, Didierjean, C. | | Deposit date: | 2014-03-13 | | Release date: | 2014-07-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural and Spectroscopic Insights into BolA-Glutaredoxin Complexes.
J.Biol.Chem., 289, 2014
|
|
2MM9
 
 | | Solution structure of reduced BolA2 from Arabidopsis thaliana | | Descriptor: | BolA2 | | Authors: | Roret, T, Tsan, P, Couturier, J, Rouhier, N, Didierjean, C. | | Deposit date: | 2014-03-13 | | Release date: | 2014-07-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural and Spectroscopic Insights into BolA-Glutaredoxin Complexes.
J.Biol.Chem., 289, 2014
|
|
5LKD
 
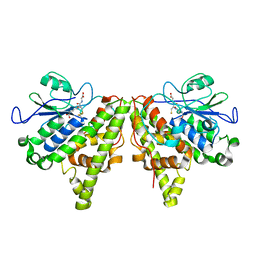 | | Crystal structure of the Xi glutathione transferase ECM4 from Saccharomyces cerevisiae in complex with glutathione | | Descriptor: | GLUTATHIONE, Glutathione S-transferase omega-like 2 | | Authors: | Schwartz, M, Didierjean, C, Hecker, A, Girardet, J.M, Morel-Rouhier, M, Gelhaye, E, Favier, F. | | Deposit date: | 2016-07-22 | | Release date: | 2016-10-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal Structure of Saccharomyces cerevisiae ECM4, a Xi-Class Glutathione Transferase that Reacts with Glutathionyl-(hydro)quinones.
Plos One, 11, 2016
|
|
5LKB
 
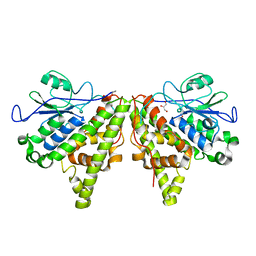 | | Crystal structure of the Xi glutathione transferase ECM4 from Saccharomyces cerevisiae | | Descriptor: | GLYCEROL, Glutathione S-transferase omega-like 2 | | Authors: | Schwartz, M, Didierjean, C, Hecker, A, Girardet, J.M, Morel-Rouhier, M, Gelhaye, E, Favier, F. | | Deposit date: | 2016-07-22 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structure of Saccharomyces cerevisiae ECM4, a Xi-Class Glutathione Transferase that Reacts with Glutathionyl-(hydro)quinones.
Plos One, 11, 2016
|
|
3S1J
 
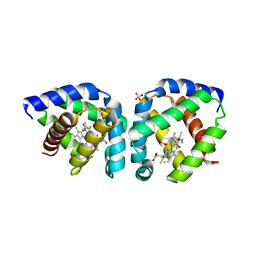 | | Crystal structure of acetate-bound hell's gate globin I | | Descriptor: | ACETATE ION, Hemoglobin-like flavoprotein, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Teh, A.H, Saito, J.A, Baharuddin, A, Tuckerman, J.R, Newhouse, J.S, Kanbe, M, Newhouse, E.I, Rahim, R.A, Favier, F, Didierjean, C, Sousa, E.H.S, Stott, M.B, Dunfield, P.F, Gonzalez, G, Gilles-Gonzalez, M.A, Najimudin, N, Alam, M. | | Deposit date: | 2011-05-15 | | Release date: | 2011-09-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Hell's Gate globin I: an acid and thermostable bacterial hemoglobin resembling mammalian neuroglobin
Febs Lett., 585, 2011
|
|
3S1I
 
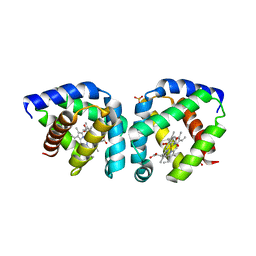 | | Crystal structure of oxygen-bound hell's gate globin I | | Descriptor: | Hemoglobin-like flavoprotein, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Teh, A.H, Saito, J.A, Baharuddin, A, Tuckerman, J.R, Newhouse, J.S, Kanbe, M, Newhouse, E.I, Rahim, R.A, Favier, F, Didierjean, C, Sousa, E.H.S, Stott, M.B, Dunfield, P.F, Gonzalez, G, Gilles-Gonzalez, M.A, Najimudin, N, Alam, M. | | Deposit date: | 2011-05-15 | | Release date: | 2011-09-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Hell's Gate globin I: an acid and thermostable bacterial hemoglobin resembling mammalian neuroglobin
Febs Lett., 585, 2011
|
|
6SR9
 
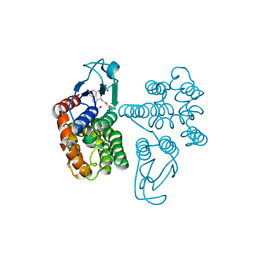 | | Crystal structure of glutathione transferase Omega 2C from Trametes versicolor in complex with oxyresveratrol | | Descriptor: | GLUTATHIONE, Uncharacterized protein, trans-oxyresveratrol | | Authors: | Schwartz, M, Favier, F, Didierjean, C. | | Deposit date: | 2019-09-05 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.845 Å) | | Cite: | Diversity of Omega Glutathione Transferases in mushroom-forming fungi revealed by phylogenetic, transcriptomic, biochemical and structural approaches.
Fungal Genet Biol., 148, 2021
|
|
6SRB
 
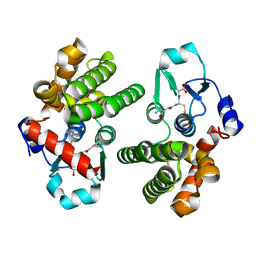 | | Crystal structure of glutathione transferase Omega 3C from Trametes versicolor | | Descriptor: | GLUTATHIONE, Uncharacterized protein | | Authors: | Schwartz, M, Favier, F, Didierjean, C. | | Deposit date: | 2019-09-05 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Diversity of Omega Glutathione Transferases in mushroom-forming fungi revealed by phylogenetic, transcriptomic, biochemical and structural approaches.
Fungal Genet Biol., 148, 2021
|
|
6SRA
 
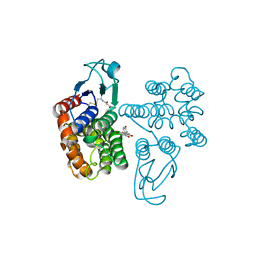 | | Crystal structure of glutathione transferase Omega 2C from Trametes versicolor in complex with naringenin | | Descriptor: | GLUTATHIONE, R-naringenin, glutathione transferase | | Authors: | Schwartz, M, Favier, F, Didierjean, C. | | Deposit date: | 2019-09-05 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Diversity of Omega Glutathione Transferases in mushroom-forming fungi revealed by phylogenetic, transcriptomic, biochemical and structural approaches.
Fungal Genet Biol., 148, 2021
|
|
6SR8
 
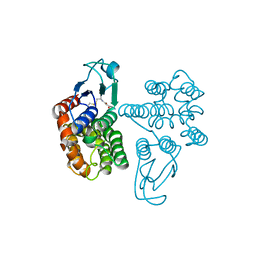 | | Crystal structure of glutathione transferase Omega 2C from Trametes versicolor | | Descriptor: | GLUTATHIONE, Uncharacterized protein | | Authors: | Schwartz, M, Favier, F, Didierjean, C. | | Deposit date: | 2019-09-05 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Diversity of Omega Glutathione Transferases in mushroom-forming fungi revealed by phylogenetic, transcriptomic, biochemical and structural approaches.
Fungal Genet Biol., 148, 2021
|
|
3RHB
 
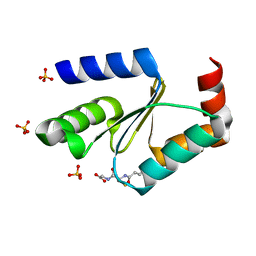 | | Crystal structure of the apo form of glutaredoxin C5 from Arabidopsis thaliana | | Descriptor: | GLUTATHIONE, Glutaredoxin-C5, chloroplastic, ... | | Authors: | Roret, T, Couturier, J, Tsan, P, Jacquot, J.P, Rouhier, N, Didierjean, C. | | Deposit date: | 2011-04-11 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Arabidopsis chloroplastic glutaredoxin c5 as a model to explore molecular determinants for iron-sulfur cluster binding into glutaredoxins.
J.Biol.Chem., 286, 2011
|
|
3RHC
 
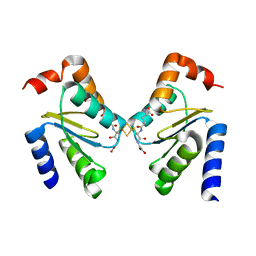 | | Crystal structure of the holo form of glutaredoxin C5 from Arabidopsis thaliana | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, GLUTATHIONE, Glutaredoxin-C5, ... | | Authors: | Roret, T, Couturier, J, Tsan, P, Jacquot, J.P, Rouhier, N, Didierjean, C. | | Deposit date: | 2011-04-11 | | Release date: | 2011-06-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Arabidopsis chloroplastic glutaredoxin c5 as a model to explore molecular determinants for iron-sulfur cluster binding into glutaredoxins.
J.Biol.Chem., 286, 2011
|
|
6GIC
 
 | |
6HPE
 
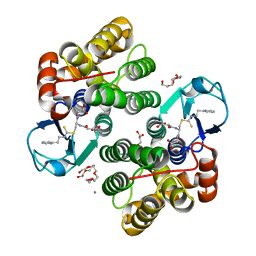 | | Crystal structure of glutathione transferase Omega 3S from Trametes versicolor in complex with the glutathione adduct of phenethyl-isothiocyanate | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Schwartz, M, Favier, F, Didierjean, C. | | Deposit date: | 2018-09-20 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The structure of Trametes versicolor glutathione transferase Omega 3S bound to its conjugation product glutathionyl-phenethylthiocarbamate reveals plasticity of its active site.
Protein Sci., 28, 2019
|
|
