1EE1
 
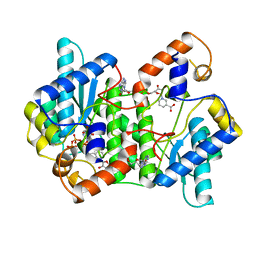 | | CRYSTAL STRUCTURE OF NH3-DEPENDENT NAD+ SYNTHETASE FROM BACILLUS SUBTILIS COMPLEXED WITH ONE MOLECULE ATP, TWO MOLECULES DEAMIDO-NAD+ AND ONE MG2+ ION | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, NH(3)-DEPENDENT NAD(+) SYNTHETASE, ... | | Authors: | Devedjiev, Y, Symersky, J, Singh, R, Jedrzejas, M, Brouillette, C, Brouillette, W, Muccio, D, Chattopadhyay, D, Delucas, L. | | Deposit date: | 2000-01-28 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Stabilization of active-site loops in NH3-dependent NAD+ synthetase from Bacillus subtilis.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1FYD
 
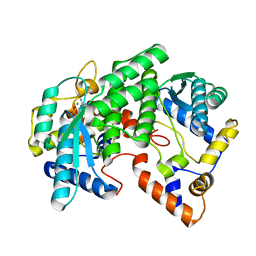 | | CRYSTAL STRUCTURE OF NH3-DEPENDENT NAD+ SYNTHETASE FROM BACILLUS SUBTILIS COMPLEXED WITH ONE MOLECULE AMP, ONE PYROPHOSPHATE ION AND ONE MG2+ ION | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, NH(3)-DEPENDENT NAD(+) SYNTHETASE, ... | | Authors: | Devedjiev, Y, Symersky, J, Singh, R, Brouillette, W, Muccio, D, Jedrzejas, M, Brouillette, C, DeLucas, L. | | Deposit date: | 2000-09-28 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Stabilization of active-site loops in NH3-dependent NAD+ synthetase from Bacillus subtilis.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1FJ2
 
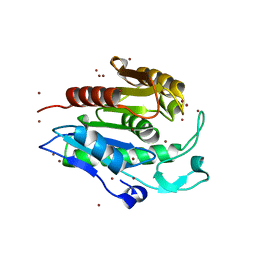 | | Crystal structure of the human acyl protein thioesterase 1 at 1.5 A resolution | | Descriptor: | BROMIDE ION, PROTEIN (ACYL PROTEIN THIOESTERASE 1) | | Authors: | Devedjiev, Y, Dauter, Z, Kuznetsov, S, Jones, T, Derewenda, Z. | | Deposit date: | 2000-08-07 | | Release date: | 2000-11-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the human acyl protein thioesterase I from a single X-ray data set to 1.5 A.
Structure Fold.Des., 8, 2000
|
|
1VPI
 
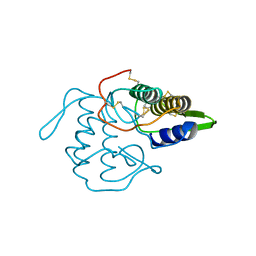 | | PHOSPHOLIPASE A2 INHIBITOR FROM VIPOXIN | | Descriptor: | PHOSPHOLIPASE A2 INHIBITOR | | Authors: | Devedjiev, Y.D, Popov, A.N. | | Deposit date: | 1996-12-17 | | Release date: | 1997-12-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | X-ray structure at 1.76 A resolution of a polypeptide phospholipase A2 inhibitor.
J.Mol.Biol., 266, 1997
|
|
1TBU
 
 | |
1IH8
 
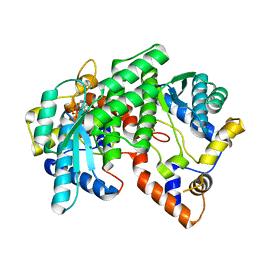 | | NH3-dependent NAD+ Synthetase from Bacillus subtilis Complexed with AMP-CPP and Mg2+ ions. | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MAGNESIUM ION, NH(3)-DEPENDENT NAD(+) synthetase | | Authors: | Devedjiev, Y, Symersky, J, Singh, R, Jedrzejas, M, Brouillette, C, Brouillette, W, Muccio, D, Chattopadhyay, D, DeLucas, L. | | Deposit date: | 2001-04-18 | | Release date: | 2001-06-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Stabilization of active-site loops in NH3-dependent NAD+ synthetase from Bacillus subtilis.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1IFX
 
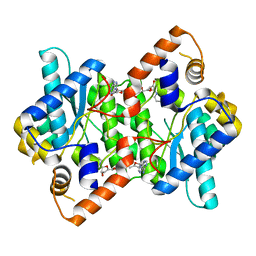 | | CRYSTAL STRUCTURE OF NH3-DEPENDENT NAD+ SYNTHETASE FROM BACILLUS SUBTILIS COMPLEXED WITH TWO MOLECULES DEAMIDO-NAD | | Descriptor: | NH(3)-DEPENDENT NAD(+) SYNTHETASE, NICOTINIC ACID ADENINE DINUCLEOTIDE | | Authors: | Devedjiev, Y, Symersky, J, Singh, R, Brouillette, W, Muccio, D, Jedrzejas, M, Brouillette, C, DeLucas, L. | | Deposit date: | 2001-04-13 | | Release date: | 2001-06-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Stabilization of active-site loops in NH3-dependent NAD+ synthetase from Bacillus subtilis.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1SBR
 
 | | The structure and function of B. subtilis YkoF gene product: the complex with thiamin | | Descriptor: | 3-(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-5-(2-HYDROXY-ETHYL)-4-METHYL-THIAZOL-3-IUM, CALCIUM ION, ykoF | | Authors: | Devedjiev, Y, Surendranath, Y, Derewenda, U, Derewenda, Z.S. | | Deposit date: | 2004-02-11 | | Release date: | 2004-10-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Structure and Ligand Binding Properties of the B.subtilis YkoF Gene Product, a Member of a Novel Family of Thiamin/HMP-binding Proteins
J.Mol.Biol., 343, 2004
|
|
1S99
 
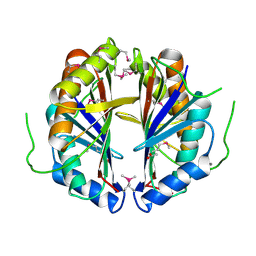 | | The structure and function of B. subtilis YkoF gene product: ligand free protein | | Descriptor: | ACETATE ION, CALCIUM ION, ykoF | | Authors: | Devedjiev, Y, Surendranath, Y, Derewenda, U, Derewenda, Z.S. | | Deposit date: | 2004-02-04 | | Release date: | 2004-10-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Structure and Ligand Binding Properties of the B.subtilis YkoF Gene Product, a Member of a Novel Family of Thiamin/HMP-binding Proteins
J.Mol.Biol., 343, 2004
|
|
2PNR
 
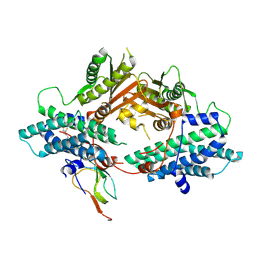 | | Crystal Structure of the asymmetric Pdk3-l2 Complex | | Descriptor: | DIHYDROLIPOIC ACID, Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, [Pyruvate dehydrogenase [lipoamide]] kinase isozyme 3 | | Authors: | Vassylyev, D.G, Steussy, C.N, Devedjiev, Y. | | Deposit date: | 2007-04-25 | | Release date: | 2007-08-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of an Asymmetric complex of Pyruvate Dehydrogenase
Kinase 3 with Lipoyl domain 2 and its Biological Implications
J.Mol.Biol., 370, 2007
|
|
5DDI
 
 | |
5TSO
 
 | |
2BJO
 
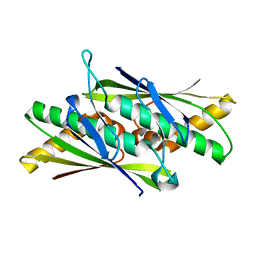 | | Crystal Structure of the Organic Hydroperoxide Resistance Protein OhrB of Bacillus subtilis | | Descriptor: | ORGANIC HYDROPEROXIDE RESISTANCE PROTEIN OHRB | | Authors: | Cooper, D.R, Surendranath, Y, Bielnicki, J, Devedjiev, Y, Joachimiak, A, Derewenda, Z.S. | | Deposit date: | 2005-02-04 | | Release date: | 2006-03-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the Bacillus Subtilis Ohrb Hydroperoxide-Resistance Protein in a Fully Oxidized State.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
1KQP
 
 | | NH3-DEPENDENT NAD+ SYNTHETASE FROM BACILLUS SUBTILIS AT 1 A RESOLUTION | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, NH(3)-dependent NAD(+) synthetase, ... | | Authors: | Symersky, J, Devedjiev, Y, Moore, K, Brouillette, C, DeLucas, L. | | Deposit date: | 2002-01-07 | | Release date: | 2002-06-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | NH3-dependent NAD+ synthetase from Bacillus subtilis at 1 A resolution.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1R6F
 
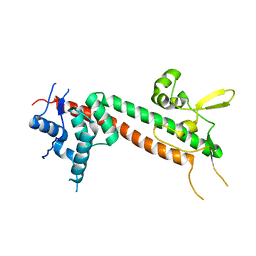 | | The structure of Yersinia pestis V-antigen, an essential virulence factor and mediator of immunity against plague | | Descriptor: | Virulence-associated V antigen | | Authors: | Derewenda, U, Mateja, A, Devedjiev, Y, Routzahn, K.M, Evdokimov, A.G, Derewenda, Z.S, Waugh, D.S. | | Deposit date: | 2003-10-15 | | Release date: | 2004-03-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | The structure of Yersinia pestis V-antigen, an essential virulence factor and mediator of immunity against plague
Structure, 12, 2004
|
|
1R6J
 
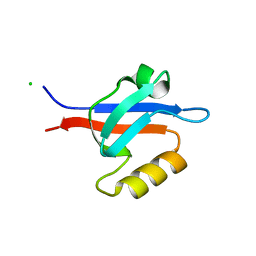 | | Ultrahigh resolution Crystal Structure of syntenin PDZ2 | | Descriptor: | CHLORIDE ION, Syntenin 1 | | Authors: | Kang, B.S, Devedjiev, Y, Derewenda, U, Derewenda, Z.S. | | Deposit date: | 2003-10-15 | | Release date: | 2004-05-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (0.73 Å) | | Cite: | The PDZ2 domain of syntenin at ultra-high resolution: bridging the gap between small molecule and macromolecular crystal chemistry
J.Mol.Biol., 338, 2004
|
|
1QVY
 
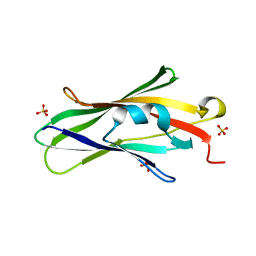 | | Crystal structure of RhoGDI K(199,200)R double mutant | | Descriptor: | Rho GDP-dissociation inhibitor 1, SULFATE ION | | Authors: | Czepas, J, Devedjiev, Y, Krowarsh, D, Derewenda, U, Derewenda, Z.S. | | Deposit date: | 2003-08-29 | | Release date: | 2004-02-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The impact of Lys-->Arg surface mutations on the crystallization of the globular domain of RhoGDI.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1H4R
 
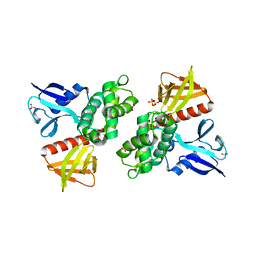 | | Crystal Structure of the FERM domain of Merlin, the Neurofibromatosis 2 Tumor Suppressor Protein. | | Descriptor: | MERLIN, SULFATE ION | | Authors: | Cooper, D.R, Kang, B.S, Sheffield, P, Devedjiev, Y, Derewenda, Z.S. | | Deposit date: | 2001-05-14 | | Release date: | 2002-01-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structure of the Ferm Domain of Merlin, the Neurofibromatosis Type 2 Gene Product.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1KMT
 
 | | Crystal structure of RhoGDI Glu(154,155)Ala mutant | | Descriptor: | Rho GDP-dissociation inhibitor 1 | | Authors: | Mateja, A, Devedjiev, Y, Krowarsh, D, Longenecker, K, Dauter, Z, Otlewski, J, Derewenda, Z.S. | | Deposit date: | 2001-12-17 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The impact of Glu-->Ala and Glu-->Asp mutations on the crystallization properties of RhoGDI: the structure of RhoGDI at 1.3 A resolution.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1OBZ
 
 | | Crystal structure of the complex of the PDZ tandem of syntenin with an interleukin 5 receptor alpha peptide. | | Descriptor: | ACETATE ION, INTERLEUKIN 5 RECEPTOR ALPHA, SYNTENIN 1 | | Authors: | Kang, B.S, Cooper, D.R, Devedjiev, Y, Derewenda, U, Derewenda, Z.S. | | Deposit date: | 2003-01-31 | | Release date: | 2003-07-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular Roots of Degenerate Specificity in Syntenin'S Pdz2 Domain: Reassessment of the Pdz Recognition Paradigm
Structure, 11, 2003
|
|
1N99
 
 | | CRYSTAL STRUCTURE OF THE PDZ TANDEM OF HUMAN SYNTENIN | | Descriptor: | Syntenin 1 | | Authors: | Kang, B.S, Cooper, D.R, Jelen, F, Devedjiev, Y, Derewenda, U, Dauter, Z, Otlewski, J, Derewenda, Z.S. | | Deposit date: | 2002-11-22 | | Release date: | 2003-04-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | PDZ Tandem of Human Syntenin: Crystal Structure and Functional Properties
Structure, 11, 2003
|
|
1NTE
 
 | | CRYSTAL STRUCTURE ANALYSIS OF THE SECOND PDZ DOMAIN OF SYNTENIN | | Descriptor: | OXYGEN ATOM, Syntenin 1 | | Authors: | Kang, B.S, Cooper, D.R, Devedjiev, Y, Derewenda, U, Derewenda, Z.S. | | Deposit date: | 2003-01-29 | | Release date: | 2003-07-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Molecular roots of degenerate specificity in syntenin's PDZ2 domain: reassessment of the PDZ recognition paradigm.
Structure, 11, 2003
|
|
1OBY
 
 | | Crystal structure of the complex of PDZ2 of syntenin with a syndecan-4 peptide. | | Descriptor: | SULFATE ION, SYNDECAN-4, SYNTENIN 1 | | Authors: | Kang, B.S, Cooper, D.R, Devedjiev, Y, Derewenda, U, Derewenda, Z.S. | | Deposit date: | 2003-01-31 | | Release date: | 2003-07-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular Roots of Degenerate Specificity in Syntenin'S Pdz2 Domain: Reassessment of the Pdz Recognition Paradigm
Structure, 11, 2003
|
|
1KLL
 
 | | Molecular basis of mitomycin C resictance in streptomyces: Crystal structures of the MRD protein with and without a drug derivative | | Descriptor: | 1,2-CIS-1-HYDROXY-2,7-DIAMINO-MITOSENE, mitomycin-binding protein | | Authors: | Martin, T.W, Dauter, Z, Devedjiev, Y, Sheffield, P, Jelen, F, He, M, Sherman, D, Otlewski, J, Derewenda, Z.S, Derewenda, U. | | Deposit date: | 2001-12-12 | | Release date: | 2002-07-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular basis of mitomycin C resistance in streptomyces: structure and function of the MRD protein.
Structure, 10, 2002
|
|
1OBX
 
 | | Crystal structure of the complex of PDZ2 of syntenin with an interleukin 5 receptor alpha peptide. | | Descriptor: | COBALT (II) ION, INTERLEUKIN 5 RECEPTOR ALPHA, SULFATE ION, ... | | Authors: | Kang, B.S, Cooper, D.R, Devedjiev, Y, Derewenda, U, Derewenda, Z.S. | | Deposit date: | 2003-01-31 | | Release date: | 2003-07-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Molecular Roots of Degenerate Specificity in Syntenin'S Pdz2 Domain: Reassessment of the Pdz Recognition Paradigm
Structure, 11, 2003
|
|
