8GHX
 
 | | Crystal Structure of CelD Cellulase from the Anaerobic Fungus Piromyces finnis | | Descriptor: | 1,2-ETHANEDIOL, Cellulase CelD | | Authors: | Dementieve, A, Kim, Y, Jedrzejczak, R, Michalska, K, Joachimiak, A. | | Deposit date: | 2023-03-13 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structure and enzymatic characterization of CelD endoglucanase from the anaerobic fungus Piromyces finnis.
Appl.Microbiol.Biotechnol., 107, 2023
|
|
8GHY
 
 | | Crystal Structure of the E154D mutant CelD Cellulase from the Anaerobic Fungus Piromyces finnis in the complex with cellotriose. | | Descriptor: | Cellulase CelD, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Dementieve, A, Kim, Y, Jedrzejczak, R, Michalska, K, Joachimiak, A. | | Deposit date: | 2023-03-13 | | Release date: | 2023-05-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and enzymatic characterization of CelD endoglucanase from the anaerobic fungus Piromyces finnis.
Appl.Microbiol.Biotechnol., 107, 2023
|
|
6W0P
 
 | | Putative kojibiose phosphorylase from human microbiome | | Descriptor: | Kojibiose phosphorylase | | Authors: | Dementiev, A, Osipiuk, J, Endres, M, Wakatsuki, S, Hess, M, Joachimiak, A. | | Deposit date: | 2020-03-02 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Putative kojibiose phosphorylase from human microbiome
to be published
|
|
8U0Q
 
 | | Co-crystal structure of optimized analog TDI-13537 provided new insights into the potency determinants of the sulfonamide inhibitor series | | Descriptor: | Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Dementiev, A.A, Michino, M, Vendome, J, Ginn, J, Bryk, R, Olland, A. | | Deposit date: | 2023-08-29 | | Release date: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Shape-Based Virtual Screening of a Billion-Compound Library Identifies Mycobacterial Lipoamide Dehydrogenase Inhibitors.
Acs Bio Med Chem Au, 3, 2023
|
|
3H5C
 
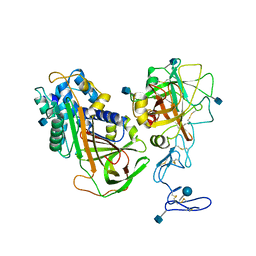 | | X-Ray Structure of Protein Z-Protein Z Inhibitor Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Protein Z-dependent protease inhibitor, Vitamin K-dependent protein Z, ... | | Authors: | Dementiev, A.A, Huang, X, Olson, S.T, Gettins, P.G.W. | | Deposit date: | 2009-04-21 | | Release date: | 2010-04-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.26 Å) | | Cite: | Basis for the specificity and activation of the serpin protein Z-dependent proteinase inhibitor (ZPI) as an inhibitor of membrane-associated factor Xa.
J.Biol.Chem., 285, 2010
|
|
6MHM
 
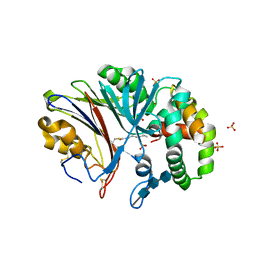 | | Crystal structure of human acid ceramidase in covalent complex with carmofur | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dementiev, A, Joachimiak, A, Doan, N. | | Deposit date: | 2018-09-18 | | Release date: | 2019-01-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.743 Å) | | Cite: | Molecular Mechanism of Inhibition of Acid Ceramidase by Carmofur.
J. Med. Chem., 62, 2019
|
|
2D26
 
 | |
6B77
 
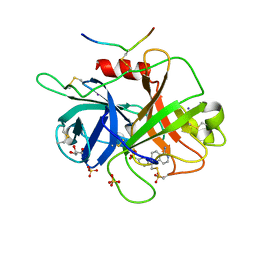 | | Structures of the two-chain human plasma factor XIIa co-crystallized with potent inhibitors | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Coagulation factor XII, GLYCEROL, ... | | Authors: | Dementiev, A.A, Silva, A, Yee, C, Flavin, M.T, Partridge, J.R. | | Deposit date: | 2017-10-03 | | Release date: | 2018-03-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structures of human plasma beta-factor XIIa cocrystallized with potent inhibitors.
Blood Adv, 2, 2018
|
|
6B74
 
 | |
6CB0
 
 | | Crystal Structure of the FAK FERM domain | | Descriptor: | Focal adhesion kinase 1 | | Authors: | Dementiev, A, Marlowe, T. | | Deposit date: | 2018-02-01 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | High resolution crystal structure of the FAK FERM domain reveals new insights on the Druggability of tyrosine 397 and the Src SH3 binding site.
BMC Mol Cell Biol, 20, 2019
|
|
1OO8
 
 | |
1OPH
 
 | | NON-COVALENT COMPLEX BETWEEN ALPHA-1-PI-PITTSBURGH AND S195A TRYPSIN | | Descriptor: | Alpha-1-antitrypsin precursor, Trypsinogen, cationic precursor | | Authors: | Dementiev, A, Simonovic, M, Volz, K, Gettins, P.G. | | Deposit date: | 2003-03-05 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Canonical inhibitor-like interactions explain reactivity of alpha1-proteinase inhibitor Pittsburgh and antithrombin with proteinases
J.Biol.Chem., 278, 2003
|
|
1SR5
 
 | | ANTITHROMBIN-ANHYDROTHROMBIN-HEPARIN TERNARY COMPLEX STRUCTURE | | Descriptor: | 2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-deoxy-2,3,6-tri-O-methyl-alpha-D-xylo-hexopyranose-(1-4)-2,3,6-tri-O-methyl-beta-D-glucopyranose-(1-4)-2,3-di-O-methyl-6-O-sulfo-alpha-D-glucopyranose-(1-4)-(2R,3R,4S,5S,6S)-6-(dihydroxymethyl)-3,4-dimethoxytetrahydro-2H-pyran-2,5-diol-(1-4)-2,3,6-tri-O-sulfo-alpha-D-glucopyranose-(1-4)-(2R,3R,4S,5S,6R)-6-(dihydroxymethyl)-3,4-dimethoxytetrahydro-2H-pyran-2,5-diol-(1-4)-1,5-anhydro-3-O-methyl-2,6-di-O-sulfo-D-glucitol, ... | | Authors: | Dementiev, A, Petitou, M, Gettins, P.G. | | Deposit date: | 2004-03-22 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The ternary complex of antithrombin-anhydrothrombin-heparin reveals the basis of inhibitor specificity.
Nat.Struct.Mol.Biol., 11, 2004
|
|
7T85
 
 | | Crystal Structure of the N-terminal Domain of the Phosphate Acetyltransferase from Escherichia coli | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Phosphate acetyltransferase, ... | | Authors: | Kim, Y, Dementiev, A, Welk, L, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-12-15 | | Release date: | 2021-12-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the N-terminal Domain of the Phosphate Acetyltransferase from Escherichia coli
To Be Published
|
|
7T88
 
 | | Crystal Structure of the C-terminal Domain of the Phosphate Acetyltransferase from Escherichia coli | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, IODIDE ION, ... | | Authors: | Kim, Y, Dementiev, A, Welk, L, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-12-15 | | Release date: | 2021-12-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of c from Escherichia coli
To Be Published
|
|
