7ODX
 
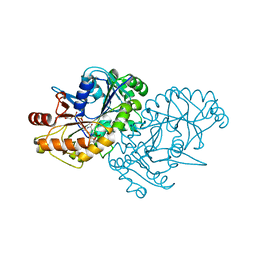 | |
1EQR
 
 | | CRYSTAL STRUCTURE OF FREE ASPARTYL-TRNA SYNTHETASE FROM ESCHERICHIA COLI | | Descriptor: | ASPARTYL-TRNA SYNTHETASE, MAGNESIUM ION | | Authors: | Rees, B, Webster, G, Delarue, M, Boeglin, M, Moras, D. | | Deposit date: | 2000-04-06 | | Release date: | 2000-06-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Aspartyl tRNA-synthetase from Escherichia coli: flexibility and adaptability to the substrates.
J.Mol.Biol., 299, 2000
|
|
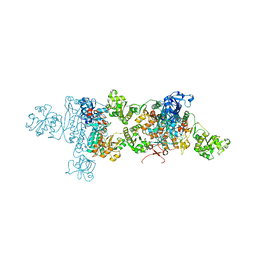
3FI0
 
 | | Crystal Structure Analysis of B. stearothermophilus Tryptophanyl-tRNA Synthetase Complexed with Tryptophan, AMP, and Inorganic Phosphate | | Descriptor: | ADENOSINE MONOPHOSPHATE, PHOSPHATE ION, TRYPTOPHAN, ... | | Authors: | Laowanapiban, P, Kapustina, M, Vonrhein, C, Delarue, M, Koehl, P, Carter Jr, C.W. | | Deposit date: | 2008-12-10 | | Release date: | 2009-02-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Independent saturation of three TrpRS subsites generates a partially assembled state similar to those observed in molecular simulations.
Proc.Natl.Acad.Sci.Usa, 106, 2009
|
|
6T8H
 
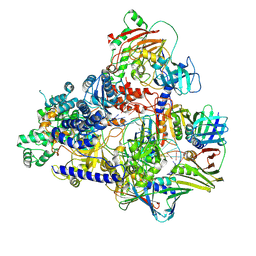 | | Cryo-EM structure of the DNA-bound PolD-PCNA processive complex from P. abyssi | | Descriptor: | DNA polymerase II small subunit, DNA polymerase sliding clamp, DNA primer, ... | | Authors: | Madru, C, Raia, P, Hugonneau Beaufet, I, Pehau-Arnaudet, G, England, P, Lindhal, E, Delarue, M, Carroni, M, Sauguet, L. | | Deposit date: | 2019-10-24 | | Release date: | 2020-03-04 | | Last modified: | 2020-04-08 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Structural basis for the increased processivity of D-family DNA polymerases in complex with PCNA.
Nat Commun, 11, 2020
|
|
6T7X
 
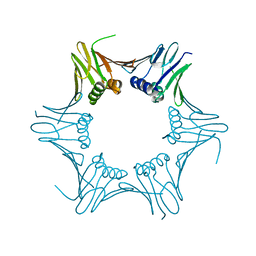 | | Crystal structure of PCNA from P. abyssi | | Descriptor: | DNA polymerase sliding clamp | | Authors: | Madru, C, Raia, P, Hugonneau Beaufet, I, Delarue, M, Carroni, M, Sauguet, L. | | Deposit date: | 2019-10-23 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the increased processivity of D-family DNA polymerases in complex with PCNA.
Nat Commun, 11, 2020
|
|
6T7Y
 
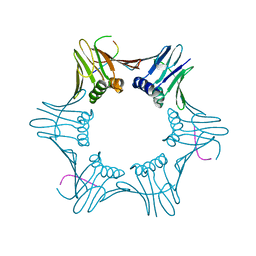 | | Structure of PCNA bound to cPIP motif of DP2 from P. abyssi | | Descriptor: | DNA polymerase sliding clamp, cPIP motif from the DP2 large subunit of PolD | | Authors: | Madru, C, Raia, P, Hugonneau Beaufet, I, Delarue, M, Carroni, M, Sauguet, L. | | Deposit date: | 2019-10-23 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the increased processivity of D-family DNA polymerases in complex with PCNA.
Nat Commun, 11, 2020
|
|
3EAM
 
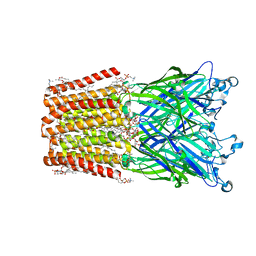 | | An open-pore structure of a bacterial pentameric ligand-gated ion channel | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, DODECYL-BETA-D-MALTOSIDE, Glr4197 protein | | Authors: | Bocquet, N, Nury, H, Baaden, M, Le Poupon, C, Changeux, J.P, Delarue, M, Corringer, P.J. | | Deposit date: | 2008-08-26 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-ray structure of a pentameric ligand-gated ion channel in an apparently open conformation.
Nature, 457, 2009
|
|
5HCJ
 
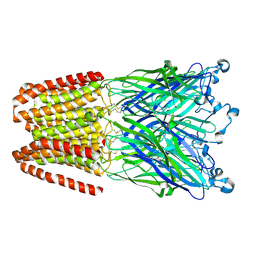 | | Cationic Ligand-Gated Ion Channel | | Descriptor: | CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, Proton-gated ion channel, ... | | Authors: | Shahsavar, A, Sauguet, L, Delarue, M. | | Deposit date: | 2016-01-04 | | Release date: | 2016-04-13 | | Last modified: | 2016-04-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Sites of Anesthetic Inhibitory Action on a Cationic Ligand-Gated Ion Channel.
Structure, 24, 2016
|
|
3FHJ
 
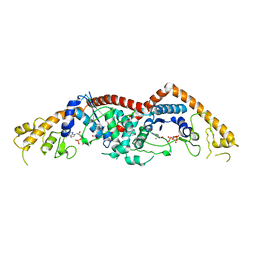 | | Independent saturation of three TrpRS subsites generates a partially-assembled state similar to those observed in molecular simulations | | Descriptor: | ADENOSINE MONOPHOSPHATE, PHOSPHATE ION, TRYPTOPHAN, ... | | Authors: | Laowanapiban, P, Kapustina, M, Vonrhein, C, Delarue, M, Koehl, P, Carter Jr, C.W. | | Deposit date: | 2008-12-09 | | Release date: | 2009-02-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Independent saturation of three TrpRS subsites generates a partially assembled state similar to those observed in molecular simulations.
Proc.Natl.Acad.Sci.Usa, 106, 2009
|
|
7B0H
 
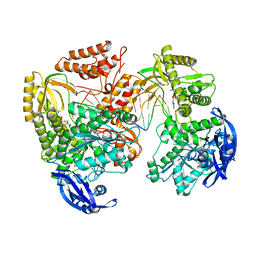 | | TgoT_6G12 Ternary complex | | Descriptor: | DNA (5'-D(P*AP*AP*CP*GP*GP*CP*AP*AP*AP*TP*GP*CP*G)-3'), DNA (5'-D(P*CP*GP*CP*AP*TP*T)-3'), DNA polymerase, ... | | Authors: | Samson, C, Legrand, P, Tekpinar, M, Rozenski, J, Abramov, M, Holliger, P, Pinheiro, V, Herdewijn, P, Delarue, M. | | Deposit date: | 2020-11-19 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural Studies of HNA Substrate Specificity in Mutants of an Archaeal DNA Polymerase Obtained by Directed Evolution.
Biomolecules, 10, 2020
|
|
7B08
 
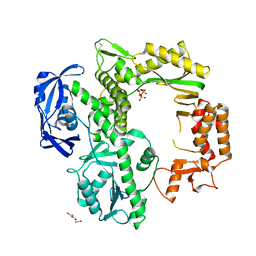 | | TgoT apo | | Descriptor: | DNA polymerase, THYMIDINE-5'-TRIPHOSPHATE, TRIETHYLENE GLYCOL | | Authors: | Samson, C, Legrand, P, Tekpinar, M, Rozenski, J, Abramov, M, Holliger, P, Pinheiro, V, Herdewijn, P, Delarue, M. | | Deposit date: | 2020-11-18 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.394 Å) | | Cite: | Structural Studies of HNA Substrate Specificity in Mutants of an Archaeal DNA Polymerase Obtained by Directed Evolution.
Biomolecules, 10, 2020
|
|
7B07
 
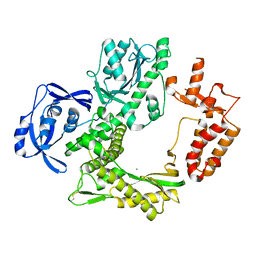 | | TgoT_6G12 apo | | Descriptor: | CALCIUM ION, DNA polymerase | | Authors: | Samson, C, Legrand, P, Tekpinar, M, Rozenski, J, Abramov, M, Holliger, P, Pinheiro, V, Herdewijn, P, Delarue, M. | | Deposit date: | 2020-11-18 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.099 Å) | | Cite: | Structural Studies of HNA Substrate Specificity in Mutants of an Archaeal DNA Polymerase Obtained by Directed Evolution.
Biomolecules, 10, 2020
|
|
7B0G
 
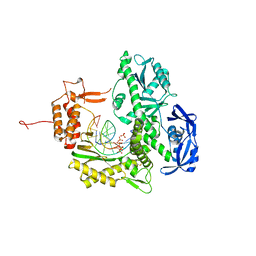 | | TgoT_6G12 binary with 2 hCTPs | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, DNA (5'-D(P*AP*TP*TP*GP*GP*CP*TP*GP*CP*CP*CP*TP*CP*C)-3'), DNA (5'-D(P*GP*GP*AP*GP*GP*GP*CP*AP*GP*()P*())-3'), ... | | Authors: | Samson, C, Legrand, P, Tekpinar, M, Rozenski, J, Abramov, M, Holliger, P, Pinheiro, V, Herdewijn, P, Delarue, M. | | Deposit date: | 2020-11-19 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Studies of HNA Substrate Specificity in Mutants of an Archaeal DNA Polymerase Obtained by Directed Evolution.
Biomolecules, 10, 2020
|
|
7B0F
 
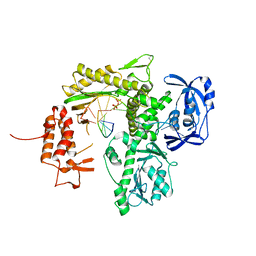 | | TgoT_6G12 Binary complex | | Descriptor: | DNA (5'-D(P*AP*AP*CP*GP*GP*CP*TP*AP*AP*TP*GP*CP*G)-3'), DNA (5'-D(P*CP*GP*CP*AP*TP*T)-3'), DNA polymerase, ... | | Authors: | Samson, C, Legrand, P, Tekpinar, M, Rozenski, J, Abramov, M, Holliger, P, Pinheiro, V, Herdewijn, P, Delarue, M. | | Deposit date: | 2020-11-19 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.797 Å) | | Cite: | Structural Studies of HNA Substrate Specificity in Mutants of an Archaeal DNA Polymerase Obtained by Directed Evolution.
Biomolecules, 10, 2020
|
|
7B06
 
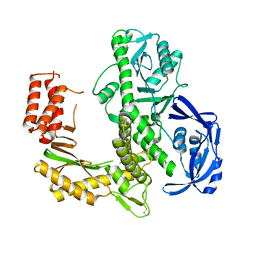 | | TgoT_RT521 apo | | Descriptor: | DNA polymerase | | Authors: | Samson, C, Legrand, P, Tekpinar, M, Rozenski, J, Abramov, M, Holliger, P, Pinheiro, V, Herdewijn, P, Delarue, M. | | Deposit date: | 2020-11-18 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.349 Å) | | Cite: | Structural Studies of HNA Substrate Specificity in Mutants of an Archaeal DNA Polymerase Obtained by Directed Evolution.
Biomolecules, 10, 2020
|
|
5HCM
 
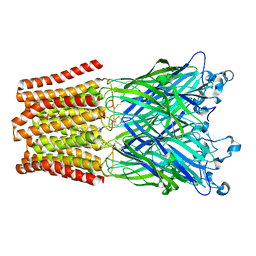 | |
3E7F
 
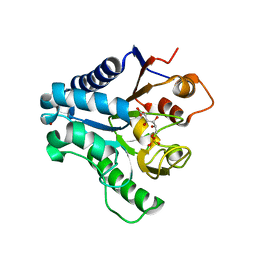 | | Crystal structure of 6-phosphogluconolactonase from Trypanosoma brucei complexed with 6-phosphogluconic acid | | Descriptor: | 6-PHOSPHOGLUCONIC ACID, 6-phosphogluconolactonase, ZINC ION | | Authors: | Poggi, L, Delarue, M, Duclert-Savatier, N, Stoven, V. | | Deposit date: | 2008-08-18 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Insights into the enzymatic mechanism of 6-phosphogluconolactonase from Trypanosoma brucei using structural data and molecular dynamics simulation.
J.Mol.Biol., 388, 2009
|
|
3EB9
 
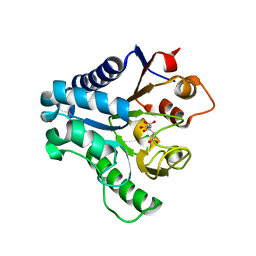 | | Crystal structure of 6-phosphogluconolactonase from trypanosoma brucei complexed with citrate | | Descriptor: | 6-phosphogluconolactonase, CITRATE ANION, ZINC ION | | Authors: | Poggi, L, Delarue, M, Duclert-Savatier, N, Stoven, V. | | Deposit date: | 2008-08-27 | | Release date: | 2009-05-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into the enzymatic mechanism of 6-phosphogluconolactonase from Trypanosoma brucei using structural data and molecular dynamics simulation.
J.Mol.Biol., 388, 2009
|
|
3LSV
 
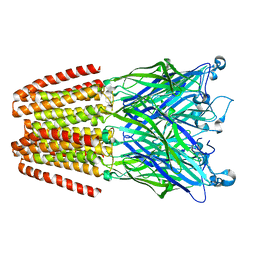 | |
1G51
 
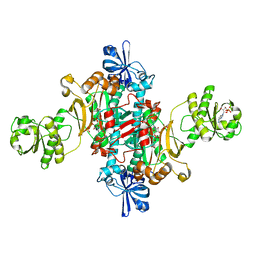 | | ASPARTYL TRNA SYNTHETASE FROM THERMUS THERMOPHILUS AT 2.4 A RESOLUTION | | Descriptor: | ADENOSINE MONOPHOSPHATE, ASPARTYL-ADENOSINE-5'-MONOPHOSPHATE, ASPARTYL-TRNA SYNTHETASE, ... | | Authors: | Poterzsman, A, Delarue, M, Thierry, J.C, Moras, D. | | Deposit date: | 2000-10-30 | | Release date: | 2000-12-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Synthesis and recognition of aspartyl-adenylate by Thermus thermophilus aspartyl-tRNA synthetase.
J.Mol.Biol., 244, 1994
|
|
3IGQ
 
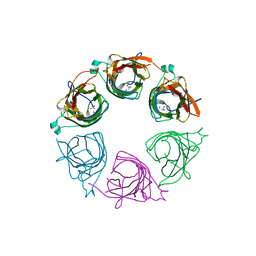 | |
6ZPA
 
 | | Cyanophage S-2L HD phosphohydrolase (DatZ) bound to dA and one catalytic Zn2+ ion | | Descriptor: | (2R,3S,5R)-5-(6-amino-9H-purin-9-yl)-tetrahydro-2-(hydroxymethyl)furan-3-ol, DatZ, LITHIUM ION, ... | | Authors: | Czernecki, D, Legrand, P, Delarue, M. | | Deposit date: | 2020-07-08 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (0.86000258 Å) | | Cite: | How cyanophage S-2L rejects adenine and incorporates 2-aminoadenine to saturate hydrogen bonding in its DNA.
Nat Commun, 12, 2021
|
|
6ZPC
 
 | | Cyanophage S-2L HD phosphohydrolase (DatZ) bound to dATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DatZ, LITHIUM ION, ... | | Authors: | Czernecki, D, Legrand, P, Delarue, M. | | Deposit date: | 2020-07-08 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.2683593 Å) | | Cite: | How cyanophage S-2L rejects adenine and incorporates 2-aminoadenine to saturate hydrogen bonding in its DNA.
Nat Commun, 12, 2021
|
|
6ZPB
 
 | | Cyanophage S-2L HD phosphohydrolase (DatZ) bound to dA and two catalytic Co2+ ions | | Descriptor: | (2R,3S,5R)-5-(6-amino-9H-purin-9-yl)-tetrahydro-2-(hydroxymethyl)furan-3-ol, COBALT (II) ION, DatZ | | Authors: | Czernecki, D, Legrand, P, Delarue, M. | | Deposit date: | 2020-07-08 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.72097385 Å) | | Cite: | How cyanophage S-2L rejects adenine and incorporates 2-aminoadenine to saturate hydrogen bonding in its DNA.
Nat Commun, 12, 2021
|
|
4ZZC
 
 | | The GLIC pentameric Ligand-Gated Ion Channel open form complexed to xenon | | Descriptor: | ACETATE ION, CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, ... | | Authors: | Sauguet, L, Fourati, Z, Prange, T, Delarue, M, Colloc'h, N. | | Deposit date: | 2015-05-22 | | Release date: | 2016-03-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis for Xenon Inhibition in a Cationic Pentameric Ligand-Gated Ion Channel.
Plos One, 11, 2016
|
|
