6QZ7
 
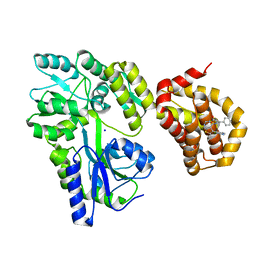 | | Structure of MBP-Mcl-1 in complex with compound 8b | | Descriptor: | (2~{R})-2-[[6-ethyl-5-(1~{H}-indol-4-yl)thieno[2,3-d]pyrimidin-4-yl]amino]-3-phenyl-propanoic acid, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, SODIUM ION, ... | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-11 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
6QXJ
 
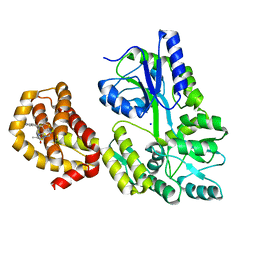 | | Structure of MBP-Mcl-1 in complex with compound 6a | | Descriptor: | (2~{R})-2-[[6-ethyl-5-(1~{H}-indol-5-yl)thieno[2,3-d]pyrimidin-4-yl]amino]propanoic acid, Maltose-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, SODIUM ION, ... | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-07 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
1MN3
 
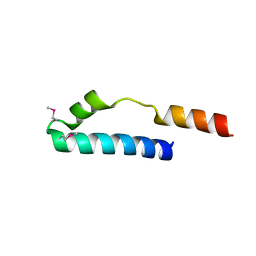 | | Cue domain of yeast Vps9p | | Descriptor: | Vacuolar protein sorting-associated protein VPS9 | | Authors: | Prag, G, Misra, S, Jones, E, Ghirlando, R, Davies, B.A, Horazdovsky, B.F, Hurley, J.H. | | Deposit date: | 2002-09-04 | | Release date: | 2003-06-10 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanism of Ubiquitin Recognition by the CUE Domain of Vps9p
Cell(Cambridge,Mass.), 113, 2003
|
|
6QGD
 
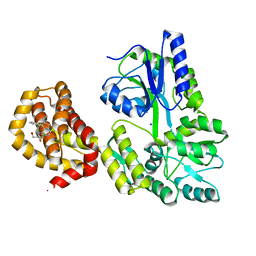 | | Structure of human Mcl-1 in complex with thienopyrimidine inhibitor | | Descriptor: | 2-[(6-ethyl-5-phenyl-thieno[2,3-d]pyrimidin-4-yl)amino]-3-oxidanyl-propanoic acid, Maltose-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, SODIUM ION, ... | | Authors: | Dokurno, P, Murray, J, Davidson, J, Chen, I, Davis, B, Graham, C.J, Harris, R, Jordan, A.M, Matassova, N, Pedder, C, Ray, S, Roughley, S, Smith, J, Walmsley, C, Wang, Y, Whitehead, N, Williamson, D.S, Casara, P, Le Diguarher, T, Hickman, J, Stark, J, Kotschy, A, Geneste, O, Hubbard, R.E. | | Deposit date: | 2019-01-11 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Establishing Drug Discovery and Identification of Hit Series for the Anti-apoptotic Proteins, Bcl-2 and Mcl-1.
Acs Omega, 4, 2019
|
|
3OB0
 
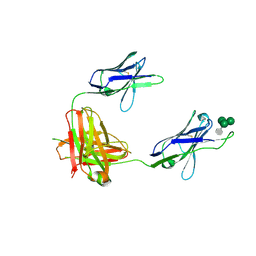 | | A non-self sugar mimic of the HIV glycan shield shows enhanced antigenicity | | Descriptor: | 7-deoxy-L-glycero-alpha-D-manno-heptopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose, Fab 2G12, heavy chain, ... | | Authors: | Doores, K.J, Fulton, Z, Hong, V, Patel, M.K, Scanlan, C.N, Wormald, M.R, Finn, M.G, Burton, D.R, Wilson, I.A, Davis, B.G. | | Deposit date: | 2010-08-06 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A nonself sugar mimic of the HIV glycan shield shows enhanced antigenicity.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1P3Q
 
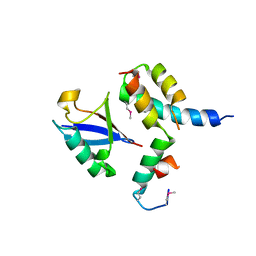 | | Mechanism of Ubiquitin Recognition by the CUE Domain of VPS9 | | Descriptor: | Ubiquitin, Vacuolar protein sorting-associated protein VPS9 | | Authors: | Prag, G, Misra, S, Jones, E.A, Ghirlando, R, Davies, B.A, Horazdovsky, B.F, Hurley, J.H. | | Deposit date: | 2003-04-18 | | Release date: | 2003-06-24 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of Ubiquitin Recognition by the CUE Domain of Vps9p.
Cell(Cambridge,Mass.), 113, 2003
|
|
1I11
 
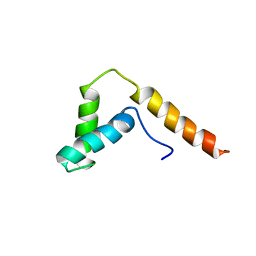 | | SOLUTION STRUCTURE OF THE DNA BINDING DOMAIN, SOX-5 HMG BOX FROM MOUSE | | Descriptor: | TRANSCRIPTION FACTOR SOX-5 | | Authors: | Cary, P.D, Read, C.M, Davis, B, Driscoll, P.C, Crane-Robinson, C. | | Deposit date: | 2001-01-30 | | Release date: | 2001-02-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of the DNA-binding domain of mouse Sox-5.
Protein Sci., 10, 2001
|
|
4YRZ
 
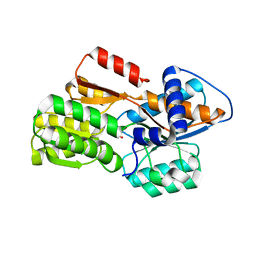 | | Crystal Structure of LuxP In Complex With L-xylulofuranose-1,2-borate | | Descriptor: | Autoinducer 2-binding periplasmic protein LuxP, L-xylulofuranose-1,2-borate | | Authors: | McDonough, M.A, Sattin, S, Gardner, P.M, Liu, C, Davis, B.G. | | Deposit date: | 2015-03-16 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.571 Å) | | Cite: | Discovery of Glycomimetic Agonists from a Protocell Metabolism, Proto-natural Product Libraries
To Be Published
|
|
4YP9
 
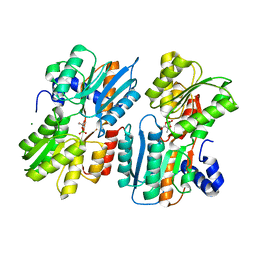 | | Crystal Structure of LuxP In Complex With a Formose Derived AI-2 Analogue | | Descriptor: | 1-deoxy-alpha-L-xylulofuranose-1,2-borate, Autoinducer 2-binding periplasmic protein LuxP, CALCIUM ION, ... | | Authors: | McDonough, M.A, Sattin, S, Gardner, P.M, Liu, C, Davis, B.G. | | Deposit date: | 2015-03-12 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of Glycomimetic Agonists from a Protocell Metabolism, Proto-natural Product Libraries
To Be Published
|
|
1BFJ
 
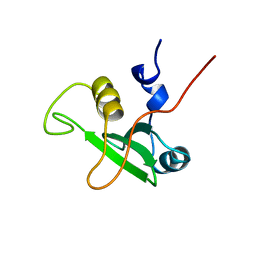 | | SOLUTION STRUCTURE OF THE C-TERMINAL SH2 DOMAIN OF THE P85ALPHA REGULATORY SUBUNIT OF PHOSPHOINOSITIDE 3-KINASE, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | P85 ALPHA | | Authors: | Siegal, G, Davis, B, Kristensen, S.M, Sankar, A, Linacre, J, Stein, R.C, Panayotou, G, Waterfield, M.D, Driscoll, P.C. | | Deposit date: | 1997-11-18 | | Release date: | 1998-02-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C-terminal SH2 domain of the p85 alpha regulatory subunit of phosphoinositide 3-kinase.
J.Mol.Biol., 276, 1998
|
|
1BFI
 
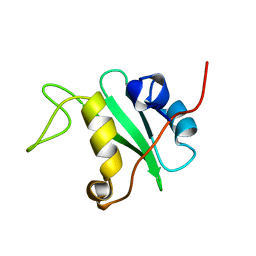 | | SOLUTION STRUCTURE OF THE C-TERMINAL SH2 DOMAIN OF THE P85ALPHA REGULATORY SUBUNIT OF PHOSPHOINOSITIDE 3-KINASE, NMR, 30 STRUCTURES | | Descriptor: | P85 ALPHA | | Authors: | Siegal, G, Davis, B, Kristensen, S.M, Sankar, A, Linacre, J, Stein, R.C, Panayotou, G, Waterfield, M.D, Driscoll, P.C. | | Deposit date: | 1997-11-18 | | Release date: | 1998-02-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C-terminal SH2 domain of the p85 alpha regulatory subunit of phosphoinositide 3-kinase.
J.Mol.Biol., 276, 1998
|
|
2YNK
 
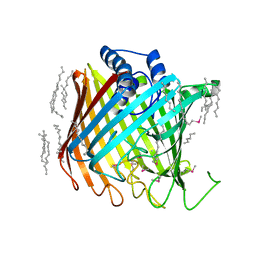 | | Wzi, an Outer Membrane Protein Involved in Group 1 Capsule Assembly in Escherichia coli, is a Carbohydrate Binding Beta-Barrel | | Descriptor: | DODECANE, HEXANE, N-OCTANE, ... | | Authors: | Bushell, S.R, Mainprize, I.L, Wear, M.A, Lou, H, Kong, L, Davis, B, Bayley, H, Whitfield, C, Naismith, J.H. | | Deposit date: | 2012-10-16 | | Release date: | 2013-05-15 | | Last modified: | 2013-05-29 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Wzi is an Outer Membrane Lectin that Underpins Group 1 Capsule Assembly in Escherichia Coli.
Structure, 21, 2013
|
|
