7T3E
 
 | | Structure of the sialic acid bound Tripartite ATP-independent Periplasmic (TRAP) periplasmic component SiaP from Photobacterium profundum | | Descriptor: | N-acetyl-beta-neuraminic acid, SULFATE ION, TRAP-type C4-dicarboxylate transport system, ... | | Authors: | Davies, J.S, Currie, M.J, North, R.A, Dobson, R.C.J. | | Deposit date: | 2021-12-07 | | Release date: | 2022-12-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Structure and mechanism of a tripartite ATP-independent periplasmic TRAP transporter.
Nat Commun, 14, 2023
|
|
8B01
 
 | | Cryo-EM structure of the Tripartite ATP-independent Periplasmic (TRAP) transporter SiaQM from Photobacterium profundum in a nanodisc | | Descriptor: | DECANE, DOCOSANE, HEXANE, ... | | Authors: | Davies, J.S, North, R.A, Dobson, R.C.J. | | Deposit date: | 2022-09-06 | | Release date: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structure and mechanism of a tripartite ATP-independent periplasmic TRAP transporter.
Nat Commun, 14, 2023
|
|
8THI
 
 | | Cryo-EM structure of the Tripartite ATP-independent Periplasmic (TRAP) transporter SiaQM from Haemophilus influenzae (parallel dimer) | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, SODIUM ION, Sialic acid TRAP transporter permease protein SiaT | | Authors: | Davies, J.S, Currie, M.C, Dobson, R.C.J, North, R.A. | | Deposit date: | 2023-07-16 | | Release date: | 2023-11-22 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Structural and biophysical analysis of a Haemophilus influenzae tripartite ATP-independent periplasmic (TRAP) transporter.
Elife, 12, 2024
|
|
8THJ
 
 | | Cryo-EM structure of the Tripartite ATP-independent Periplasmic (TRAP) transporter SiaQM from Haemophilus influenzae (antiparallel dimer) | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, PHOSPHATIDYLETHANOLAMINE, SODIUM ION, ... | | Authors: | Davies, J.S, Currie, M.C, Dobson, R.C.J, North, R.A. | | Deposit date: | 2023-07-16 | | Release date: | 2023-11-22 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Structural and biophysical analysis of a Haemophilus influenzae tripartite ATP-independent periplasmic (TRAP) transporter.
Elife, 12, 2024
|
|
3EE1
 
 | |
6Q27
 
 | | N-acetylmannosamine kinase with N-acetylmannosamine from Staphylococcus aureus | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-mannopyranose, Glucokinase | | Authors: | Coombes, D, Horne, C.R, Davies, J.S, Dobson, R.C.J. | | Deposit date: | 2019-08-07 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The basis for non-canonical ROK family function in theN-acetylmannosamine kinase from the pathogenStaphylococcus aureus.
J.Biol.Chem., 295, 2020
|
|
6O15
 
 | | Crystal structure of a putative oxidoreductase YjhC from Escherichia coli in complex with NAD(H) | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, Uncharacterized oxidoreductase YjhC | | Authors: | Horne, C.R, Kind, L, Davies, J.S, Dobson, R.C. | | Deposit date: | 2019-02-17 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | On the structure and function of Escherichia coli YjhC: An oxidoreductase involved in bacterial sialic acid metabolism.
Proteins, 88, 2020
|
|
7QHA
 
 | | Cryo-EM structure of the Tripartite ATP-independent Periplasmic (TRAP) transporter SiaQM from Photobacterium profundum in amphipol | | Descriptor: | Megabody c7HopQ, Putative TRAP-type C4-dicarboxylate transport system, large permease component, ... | | Authors: | North, R.A, Davies, J.S, Morado, D, Dobson, R.C.J. | | Deposit date: | 2021-12-11 | | Release date: | 2022-12-21 | | Last modified: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structure and mechanism of a tripartite ATP-independent periplasmic TRAP transporter.
Nat Commun, 14, 2023
|
|
1QTO
 
 | | 1.5 A CRYSTAL STRUCTURE OF A BLEOMYCIN RESISTANCE DETERMINANT FROM BLEOMYCIN-PRODUCING STREPTOMYCES VERTICILLUS | | Descriptor: | BLEOMYCIN-BINDING PROTEIN | | Authors: | Kawano, Y, Kumagai, T, Muta, K, Matoba, Y, Davies, J, Sugiyama, M. | | Deposit date: | 1999-06-28 | | Release date: | 2000-06-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The 1.5 A crystal structure of a bleomycin resistance determinant from bleomycin-producing Streptomyces verticillus.
J.Mol.Biol., 295, 2000
|
|
6E8L
 
 | | Crystal Structure of Alkyl hydroperoxidase D (AhpD) from Streptococcus pneumoniae (Strain D39/ NCTC 7466) | | Descriptor: | Alkyl hydroperoxide reductase AhpD | | Authors: | Meng, Y, Davies, J, North, R, Coombes, D, Horne, C, Hampton, M, Dobson, R. | | Deposit date: | 2018-07-30 | | Release date: | 2019-08-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-function analyses of alkylhydroperoxidase D fromStreptococcus pneumoniaereveal an unusual three-cysteine active site architecture.
J.Biol.Chem., 295, 2020
|
|
7B15
 
 | |
7B13
 
 | |
6YLU
 
 | | 14-3-3sigma in complex with BLNKpT152 phosphopeptide crystal structure | | Descriptor: | 14-3-3 protein sigma, BLNKpT152, MAGNESIUM ION | | Authors: | Soini, L, Leysen, S, Davis, J, Ottmann, C. | | Deposit date: | 2020-04-07 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | 14-3-3sigma in complex with BLNKpT152 phosphopeptide crystal structure
J.Struct.Biol., 2020
|
|
6ZCJ
 
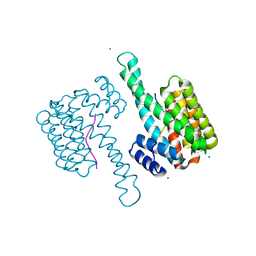 | | 14-3-3sigma in complex with SLP76pS376 phosphopeptide crystal structure | | Descriptor: | 14-3-3 protein sigma, MAGNESIUM ION, SLP76pS376 | | Authors: | Soini, L, Leysen, S, Davis, J, Ottmann, C. | | Deposit date: | 2020-06-11 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | The 14-3-3/SLP76 protein-protein interaction in T-cell receptor signalling: a structural and biophysical characterization.
Febs Lett., 595, 2021
|
|
2W3L
 
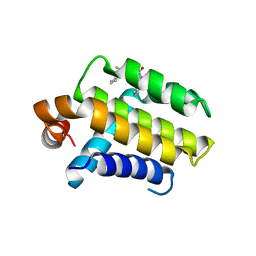 | | Crystal Structure of Chimaeric Bcl2-xL and Phenyl Tetrahydroisoquinoline Amide Complex | | Descriptor: | 1-(2-{[(3S)-3-(aminomethyl)-3,4-dihydroisoquinolin-2(1H)-yl]carbonyl}phenyl)-4-chloro-5-methyl-N,N-diphenyl-1H-pyrazole-3-carboxamide, APOPTOSIS REGULATOR BCL-2 | | Authors: | Porter, J, Payne, A, de Candole, B, Ford, D, Hutchinson, B, Trevitt, G, Turner, J, Edwards, C, Watkins, C, Whitcombe, I, Davis, J, Stubberfield, C, Fisher, M, Lamers, M. | | Deposit date: | 2008-11-13 | | Release date: | 2008-12-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Tetrahydroisoquinoline Amide Substituted Phenyl Pyrazoles as Selective Bcl-2 Inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
7JU4
 
 | | Radial spoke 2 stalk, IDAc, and N-DRC attached with doublet microtubule | | Descriptor: | 28 kDa inner dynein arm light chain, axonemal, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Gui, M, Ma, M, Sze-Tu, E, Wang, X, Koh, F, Zhong, E, Berger, B, Davis, J, Dutcher, S, Zhang, R, Brown, A. | | Deposit date: | 2020-08-19 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of radial spokes and associated complexes important for ciliary motility.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7JTK
 
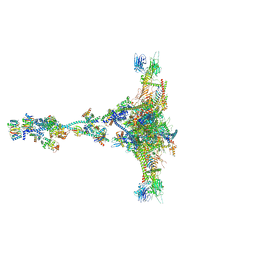 | | Radial spoke 1 isolated from Chlamydomonas reinhardtii | | Descriptor: | Cytochrome b5 heme-binding domain-containing protein, Dynein 8 kDa light chain, flagellar outer arm, ... | | Authors: | Gui, M, Ma, M, Sze-Tu, E, Wang, X, Koh, F, Zhong, E, Berger, B, Davis, J, Dutcher, S, Zhang, R, Brown, A. | | Deposit date: | 2020-08-17 | | Release date: | 2020-12-16 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of radial spokes and associated complexes important for ciliary motility.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7JTS
 
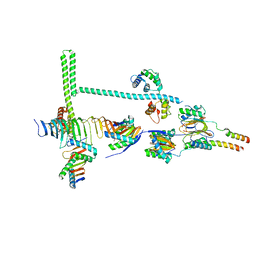 | | Stalk of radial spoke 1 attached with doublet microtubule from Chlamydomonas reinhardtii | | Descriptor: | Calmodulin, Dynein 8 kDa light chain, flagellar outer arm, ... | | Authors: | Gui, M, Ma, M, Sze-Tu, E, Wang, X, Koh, F, Zhong, E, Berger, B, Davis, J, Dutcher, S, Zhang, R, Brown, A. | | Deposit date: | 2020-08-18 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structures of radial spokes and associated complexes important for ciliary motility.
Nat.Struct.Mol.Biol., 28, 2021
|
|
1UZR
 
 | | Crystal Structure of the Class Ib Ribonucleotide Reductase R2F-2 subunit from Mycobacterium tuberculosis | | Descriptor: | CITRIC ACID, FE (III) ION, GLYCEROL, ... | | Authors: | Uppsten, M, Davis, J, Rubin, H, Uhlin, U. | | Deposit date: | 2004-03-15 | | Release date: | 2004-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Biologically Active Form of Class Ib Ribonucleotide Reductase Small Subunit from Mycobacterium Tuberculosis
FEBS Lett., 569, 2004
|
|
1OEC
 
 | | FGFr2 kinase domain | | Descriptor: | 4-ARYL-2-PHENYLAMINO PYRIMIDINE, FIBROBLAST GROWTH FACTOR RECEPTOR 2, SULFATE ION | | Authors: | Ceska, T.A, Owens, R, Doyle, C, Hamlyn, P, Crabbe, T, Moffat, D, Davis, J, Martin, R, Perry, M.J. | | Deposit date: | 2003-03-24 | | Release date: | 2004-10-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Crystal Structure of the Fgfr2 Tyrosine Kinase Domain in Complex with 4-Aryl-2-Phenylamino Pyrimidine Angiogenesis Inhibitors
To be Published
|
|
1GJO
 
 | | The FGFr2 tyrosine kinase domain | | Descriptor: | FIBROBLAST GROWTH FACTOR RECEPTOR 2, SULFATE ION | | Authors: | Ceska, T.A, Owens, R, Doyle, C, Hamlyn, P, Crabbe, T, Moffat, D, Davis, J, Martin, R, Perry, M.J. | | Deposit date: | 2001-07-31 | | Release date: | 2003-08-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Fgfr2 Tyrosine Kinase Domain
To be Published
|
|
