5ZAU
 
 | |
7EZ0
 
 | | Apo L-21 ScaI Tetrahymena ribozyme | | Descriptor: | Apo L-21 ScaI Tetrahymena ribozyme, MAGNESIUM ION | | Authors: | Su, Z, Zhang, K, Kappel, K, Luo, B, Das, R, Chiu, W. | | Deposit date: | 2021-06-01 | | Release date: | 2021-08-25 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Cryo-EM structures of full-length Tetrahymena ribozyme at 3.1 angstrom resolution.
Nature, 596, 2021
|
|
5XFU
 
 | | Domain swapped dimer crystal structure of loop1 deletion mutant in Single-chain Monellin | | Descriptor: | Monellin chain B,Monellin chain A | | Authors: | Surana, P, Nandwani, N, Udgaonkar, J, Gosavi, S, Das, R. | | Deposit date: | 2017-04-11 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.611 Å) | | Cite: | Amino-acid composition after loop deletion drives domain swapping
Protein Sci., 26, 2017
|
|
5XQM
 
 | |
5YCU
 
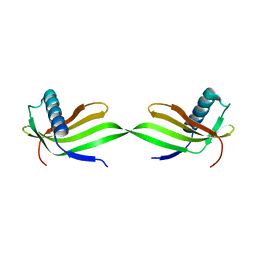 | | Domain swapped dimer of engineered hairpin loop1 mutant in Single-chain Monellin | | Descriptor: | Single chain monellin | | Authors: | Surana, P, Nandwani, N, Udgaonkar, J.B, Gosavi, S, Das, R. | | Deposit date: | 2017-09-08 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | A five-residue motif for the design of domain swapping in proteins.
Nat Commun, 10, 2019
|
|
5YCW
 
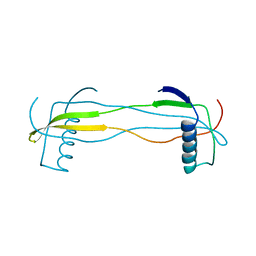 | | Double domain swapped dimer of engineered hairpin loop1 and loop3 mutant in Single-chain Monellin | | Descriptor: | single chain monellin | | Authors: | Surana, P, Nandwani, N, Udgaonkar, J.B, Gosavi, S, Das, R. | | Deposit date: | 2017-09-08 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.285 Å) | | Cite: | A five-residue motif for the design of domain swapping in proteins.
Nat Commun, 10, 2019
|
|
5YCT
 
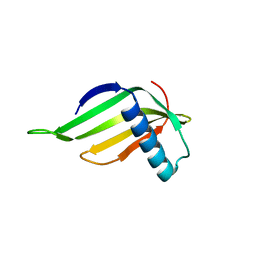 | | Engineered hairpin loop3 mutant monomer in Single-chain Monellin | | Descriptor: | Single chain Monellin | | Authors: | Surana, P, Nandwani, N, Udgaonkar, J.B, Gosavi, S, Das, R. | | Deposit date: | 2017-09-08 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | A five-residue motif for the design of domain swapping in proteins.
Nat Commun, 10, 2019
|
|
6IWJ
 
 | | A designed domain swapped dimer | | Descriptor: | Archeal Protein MK0293 | | Authors: | Nandwani, N, Negi, H, Das, R. | | Deposit date: | 2018-12-05 | | Release date: | 2019-02-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A five-residue motif for the design of domain swapping in proteins.
Nat Commun, 10, 2019
|
|
7UPH
 
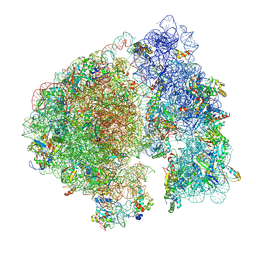 | | Structure of a ribosome with tethered subunits | | Descriptor: | 30S ribosomal protein S10, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Kim, D.S, Watkins, A, Bidstrup, E, Lee, J, Topkar, V.V, Kofman, C, Schwarz, K.J, Liu, Y, Pintilie, G, Roney, E, Das, R, Jewett, M.C. | | Deposit date: | 2022-04-15 | | Release date: | 2022-08-17 | | Last modified: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Three-dimensional structure-guided evolution of a ribosome with tethered subunits.
Nat.Chem.Biol., 18, 2022
|
|
6J4I
 
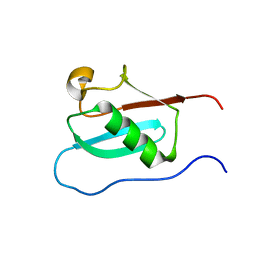 | |
6K5R
 
 | | Complex of SUMO2 with Phosphorylated viral SIM IE2 | | Descriptor: | ASP-THR-ALA-GLY-CYS-ILE-VAL-ILE-SEP-ASP-SEP-GLU, Small ubiquitin-related modifier 3 | | Authors: | Chatterjee, K.S, Das, R. | | Deposit date: | 2019-05-30 | | Release date: | 2019-08-07 | | Last modified: | 2019-12-25 | | Method: | SOLUTION NMR | | Cite: | Casein kinase-2-mediated phosphorylation increases the SUMO-dependent activity of the cytomegalovirus transactivator IE2.
J.Biol.Chem., 294, 2019
|
|
7XSN
 
 | | Native Tetrahymena ribozyme conformation | | Descriptor: | RNA (387-MER) | | Authors: | Li, S, Palo, M, Pintilie, G, Zhang, X, Su, Z, Kappel, K, Chiu, W, Zhang, K, Das, R. | | Deposit date: | 2022-05-14 | | Release date: | 2022-08-03 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Topological crossing in the misfolded Tetrahymena ribozyme resolved by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XSL
 
 | | Misfolded Tetrahymena ribozyme conformation 2 | | Descriptor: | RNA (388-MER) | | Authors: | Li, S, Palo, M, Pintilie, G, Zhang, X, Su, Z, Kappel, K, Chiu, W, Zhang, K, Das, R. | | Deposit date: | 2022-05-14 | | Release date: | 2022-08-03 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Topological crossing in the misfolded Tetrahymena ribozyme resolved by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XSM
 
 | | Misfolded Tetrahymena ribozyme conformation 3 | | Descriptor: | RNA (388-MER) | | Authors: | Li, S, Palo, M, Pintilie, G, Zhang, X, Su, Z, Kappel, K, Chiu, W, Zhang, K, Das, R. | | Deposit date: | 2022-05-14 | | Release date: | 2022-08-03 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.01 Å) | | Cite: | Topological crossing in the misfolded Tetrahymena ribozyme resolved by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XSK
 
 | | Misfolded Tetrahymena ribozyme conformation 1 | | Descriptor: | RNA (388-MER) | | Authors: | Li, S, Palo, M, Pintilie, G, Zhang, X, Su, Z, Kappel, K, Chiu, W, Zhang, K, Das, R. | | Deposit date: | 2022-05-14 | | Release date: | 2022-08-03 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Topological crossing in the misfolded Tetrahymena ribozyme resolved by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4LAD
 
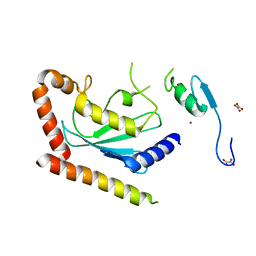 | | Crystal Structure of the Ube2g2:RING-G2BR complex | | Descriptor: | E3 ubiquitin-protein ligase AMFR, OXALATE ION, Ubiquitin-conjugating enzyme E2 G2, ... | | Authors: | Liang, Y.-H, Li, J, Das, R, Byrd, R.A, Ji, X. | | Deposit date: | 2013-06-19 | | Release date: | 2013-08-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Allosteric regulation of E2:E3 interactions promote a processive ubiquitination machine.
Embo J., 32, 2013
|
|
4L9M
 
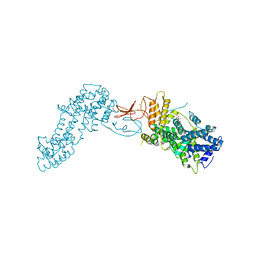 | | Autoinhibited state of the Ras-specific exchange factor RasGRP1 | | Descriptor: | CITRIC ACID, GLYCEROL, RAS guanyl-releasing protein 1, ... | | Authors: | Iwig, J.S, Vercoulen, Y, Das, R, Barros, T, Limnander, A, Che, Y, Pelton, J.G, Wemmer, D.E, Roose, J.P, Kuriyan, J. | | Deposit date: | 2013-06-18 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural analysis of autoinhibition in the Ras-specific exchange factor RasGRP1.
Elife, 2, 2013
|
|
4L9U
 
 | | Structure of C-terminal coiled coil of RasGRP1 | | Descriptor: | GLYCEROL, RAS guanyl-releasing protein 1, SULFATE ION | | Authors: | Iwig, J.S, Vercoulen, Y, Das, R, Barros, T, Limnander, A, Che, Y, Pelton, J.G, Wemmer, D.E, Roose, J.P, Kuriyan, J. | | Deposit date: | 2013-06-18 | | Release date: | 2013-08-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6014 Å) | | Cite: | Structural analysis of autoinhibition in the Ras-specific exchange factor RasGRP1.
Elife, 2, 2013
|
|
2LVQ
 
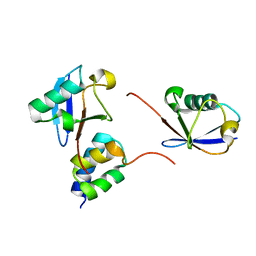 | | gp78CUE domain bound to the proximal ubiquitin of K48-linked diubiquitin | | Descriptor: | E3 ubiquitin-protein ligase AMFR, Ubiquitin | | Authors: | Liu, S, Chen, Y, Huang, T, Tarasov, S.G, King, A, Li, J, Weissman, A.M, Byrd, R.A, Das, R. | | Deposit date: | 2012-07-09 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Promiscuous Interactions of gp78 E3 Ligase CUE Domain with Polyubiquitin Chains.
Structure, 20, 2012
|
|
2NBD
 
 | |
2NBE
 
 | |
2LFX
 
 | | Structure of the duplex when (5'S)-8,5'-cyclo-2'-deoxyguanosine is placed opposite dT | | Descriptor: | DNA (5'-D(*AP*CP*AP*AP*AP*CP*AP*TP*GP*CP*AP*C)-3'), DNA (5'-D(*GP*TP*GP*CP*(2LF)P*TP*GP*TP*TP*TP*GP*T)-3') | | Authors: | Huang, H, Das, R.S, Basu, A, Stone, M.P. | | Deposit date: | 2011-07-18 | | Release date: | 2012-06-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structures of (5'S)-8,5'-Cyclo-2'-deoxyguanosine Mismatched with dA or dT.
Chem.Res.Toxicol., 25, 2012
|
|
2LFY
 
 | | Structure of the duplex when (5'S)-8,5'-cyclo-2'-deoxyguanosine is placed opposite dA | | Descriptor: | DNA (5'-D(*AP*CP*AP*AP*AP*CP*AP*AP*GP*CP*AP*C)-3'), DNA (5'-D(*GP*TP*GP*CP*(2LF)P*TP*GP*TP*TP*TP*GP*T)-3') | | Authors: | Huang, H, Das, R.S, Basu, A, Stone, M.P. | | Deposit date: | 2011-07-18 | | Release date: | 2012-06-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structures of (5'S)-8,5'-Cyclo-2'-deoxyguanosine Mismatched with dA or dT.
Chem.Res.Toxicol., 25, 2012
|
|
2LG0
 
 | | structure of the duplex containing (5'S)-8,5'-cyclo-2'-deoxyadenosine | | Descriptor: | DNA (5'-D(*AP*CP*AP*AP*AP*CP*AP*TP*GP*CP*AP*C)-3'), DNA (5'-D(*GP*TP*GP*CP*(02I)P*TP*GP*TP*TP*TP*GP*T)-3') | | Authors: | Huang, H, Das, R.S, Basu, A, Stone, M.P. | | Deposit date: | 2011-07-18 | | Release date: | 2012-06-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of (5'S)-8,5'-cyclo-2'-deoxyguanosine in DNA.
J.Am.Chem.Soc., 133, 2011
|
|
2LFA
 
 | | Oligonucleotide duplex contaning (5'S)-8,5'-cyclo-2'-deoxyguansine | | Descriptor: | DNA (5'-D(*AP*CP*AP*AP*AP*CP*AP*CP*GP*CP*AP*C)-3'), DNA (5'-D(*GP*TP*GP*CP*(2LF)P*TP*GP*TP*TP*TP*GP*T)-3') | | Authors: | Huang, H, Das, R.S, Basu, A, Stone, M.P. | | Deposit date: | 2011-06-29 | | Release date: | 2012-01-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of (5'S)-8,5'-Cyclo-2'-deoxyguanosine in DNA.
J.Am.Chem.Soc., 133, 2011
|
|
