1A17
 
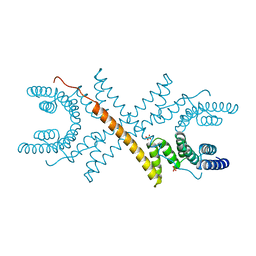 | | TETRATRICOPEPTIDE REPEATS OF PROTEIN PHOSPHATASE 5 | | Descriptor: | SERINE/THREONINE PROTEIN PHOSPHATASE 5, SULFATE ION | | Authors: | Das, A.K, Cohen, P.T.W, Barford, D. | | Deposit date: | 1997-12-23 | | Release date: | 1998-04-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The structure of the tetratricopeptide repeats of protein phosphatase 5: implications for TPR-mediated protein-protein interactions.
EMBO J., 17, 1998
|
|
1A6Q
 
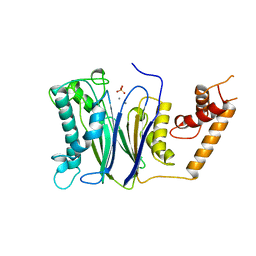 | | CRYSTAL STRUCTURE OF THE PROTEIN SERINE/THREONINE PHOSPHATASE 2C AT 2 A RESOLUTION | | Descriptor: | MANGANESE (II) ION, PHOSPHATASE 2C, PHOSPHATE ION | | Authors: | Das, A.K, Helps, N.R, Cohen, P.T.W, Barford, D. | | Deposit date: | 1998-02-27 | | Release date: | 1998-05-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the protein serine/threonine phosphatase 2C at 2.0 A resolution.
EMBO J., 15, 1996
|
|
8YRS
 
 | |
3BT4
 
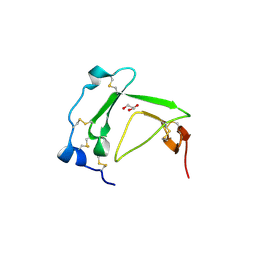 | | Crystal Structure Analysis of AmFPI-1, fungal protease inhibitor from Antheraea mylitta | | Descriptor: | Fungal protease inhibitor-1, GLYCEROL | | Authors: | Roy, S, Aravind, P, Madhurantakam, C, Ghosh, A.K, Sankarananarayanan, R, Das, A.K. | | Deposit date: | 2007-12-27 | | Release date: | 2008-12-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a fungal protease inhibitor from Antheraea mylitta
J.Struct.Biol., 166, 2009
|
|
7XLY
 
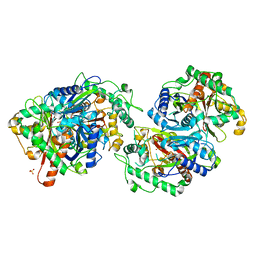 | | Crystal structure of FadA2 (Rv0243) from the fatty acid metabolic pathway of Mycobacterium tuberculosis | | Descriptor: | Probable acetyl-CoA acyltransferase FadA2 (3-ketoacyl-CoA thiolase) (Beta-ketothiolase), SULFATE ION | | Authors: | Singh, R, Kundu, P, Singh, B.K, Bhattacharyya, S, Das, A.K. | | Deposit date: | 2022-04-23 | | Release date: | 2023-04-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of FadA2 thiolase from Mycobacterium tuberculosis and prediction of its substrate specificity and membrane-anchoring properties.
Febs J., 290, 2023
|
|
7YD4
 
 | | Crystal structure of an N terminal truncated secreted protein, Rv0398c from Mycobacterium tuberculosis | | Descriptor: | GLYCEROL, Possible secreted protein | | Authors: | Saha, R, Mukherjee, S, Singh, B.K, Weiss, M.S, De, S, Das, A.K. | | Deposit date: | 2022-07-03 | | Release date: | 2023-06-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.896 Å) | | Cite: | Crystal structure of a mycobacterial secretory protein Rv0398c and in silico prediction of its export pathway.
Biochem.Biophys.Res.Commun., 672, 2023
|
|
4MY4
 
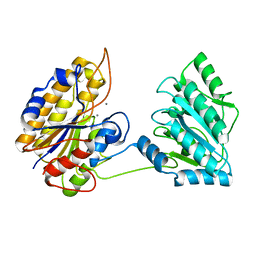 | | Crystal structure of phosphoglycerate mutase from Staphylococcus aureus. | | Descriptor: | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase, MANGANESE (II) ION | | Authors: | Roychowdhury, A, Kundu, A, Gujar, A, Bose, M, Das, A.K. | | Deposit date: | 2013-09-27 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Complete catalytic cycle of cofactor-independent phosphoglycerate mutase involves a spring-loaded mechanism
Febs J., 282, 2015
|
|
3K73
 
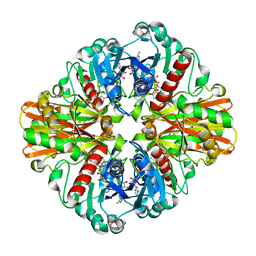 | | Crystal Structure of Phosphate bound Holo Glyceraldehyde-3-phosphate dehydrogenase 1 from MRSA252 at 2.5 Angstrom resolution | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase 1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Mukherjee, S, Dutta, D, Saha, B, Das, A.K. | | Deposit date: | 2009-10-12 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase 1 from methicillin-resistant Staphylococcus aureus MRSA252 provides novel insights into substrate binding and catalytic mechanism.
J.Mol.Biol., 401, 2010
|
|
3KSZ
 
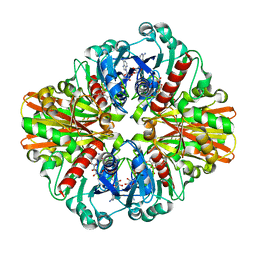 | | Crystal Structure of C151S+H178N mutant of Glyceraldehyde-3-phosphate-dehydrogenase 1 (GAPDH 1) from Staphylococcus aureus MRSA252 complexed with NAD and G3P | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, Glyceraldehyde-3-phosphate dehydrogenase 1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Mukherjee, S, Dutta, D, Saha, B, Das, A.K. | | Deposit date: | 2009-11-24 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase 1 from methicillin-resistant Staphylococcus aureus MRSA252 provides novel insights into substrate binding and catalytic mechanism.
J.Mol.Biol., 401, 2010
|
|
7YVY
 
 | |
9J1S
 
 | |
4RUV
 
 | | Crystal structure of thioredoxin 2 from Staphylococcus aureus NCTC8325 | | Descriptor: | Thioredoxin | | Authors: | Bose, M, Biswas, R, Roychowdhury, A, Bhattacharyya, S, Ghosh, A.K, Das, A.K. | | Deposit date: | 2014-11-22 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Elucidation of the mechanism of disulfide exchange between staphylococcal thioredoxin2 and thioredoxin reductase2: A structural insight.
Biochimie, 160, 2019
|
|
5F24
 
 | | Crystal structure of dual specific IMPase/NADP phosphatase bound with D-inositol-1-phosphate | | Descriptor: | CALCIUM ION, CHLORIDE ION, D-MYO-INOSITOL-1-PHOSPHATE, ... | | Authors: | Bhattacharyya, S, Dutta, D, Ghosh, A.K, Das, A.K. | | Deposit date: | 2015-12-01 | | Release date: | 2015-12-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural elucidation of the NADP(H) phosphatase activity of staphylococcal dual-specific IMPase/NADP(H) phosphatase
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5EYH
 
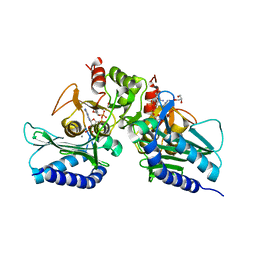 | | Crystal Structure of IMPase/NADP phosphatase complexed with NADP and Ca2+ at pH 7.0 | | Descriptor: | CALCIUM ION, GLYCEROL, Inositol monophosphatase, ... | | Authors: | Bhattacharyya, S, Dutta, D, Ghosh, A.K, Das, A.K. | | Deposit date: | 2015-11-25 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural elucidation of the NADP(H) phosphatase activity of staphylococcal dual-specific IMPase/NADP(H) phosphatase
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5EYG
 
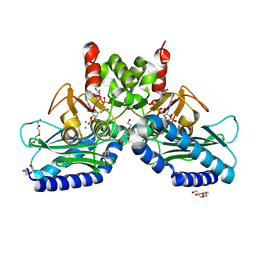 | | Crystal structure of IMPase/NADP phosphatase complexed with NADP and Ca2+ | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Bhattacharyya, S, Dutta, D, Ghosh, A.K, Das, A.K. | | Deposit date: | 2015-11-25 | | Release date: | 2015-12-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural elucidation of the NADP(H) phosphatase activity of staphylococcal dual-specific IMPase/NADP(H) phosphatase
Acta Crystallogr D Struct Biol, 72, 2016
|
|
4RV2
 
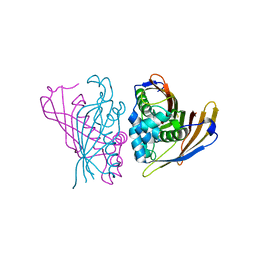 | | Crystal Structure of (3R)-hydroxyacyl-ACP dehydratase HadAB hetero-dimer from Mycobacterium smegmatis | | Descriptor: | MaoC family protein, SULFATE ION, UPF0336 protein MSMEG_1340/MSMEI_1302 | | Authors: | Biswas, R, Hazra, D, Dutta, D, Das, A.K. | | Deposit date: | 2014-11-24 | | Release date: | 2015-02-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of dehydratase component HadAB complex of mycobacterial FAS-II pathway.
Biochem.Biophys.Res.Commun., 458, 2015
|
|
5DW8
 
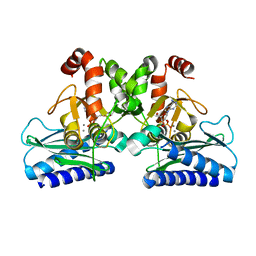 | |
5J16
 
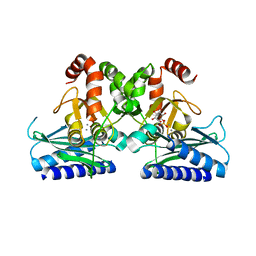 | |
7CZC
 
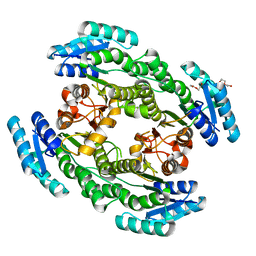 | | Crystal structure of apo-FabG from Vibrio harveyi | | Descriptor: | 3-oxoacyl-ACP reductase FabG, DI(HYDROXYETHYL)ETHER | | Authors: | Singh, B.K, Kumar, A, Paul, B, Biswas, R, Das, A.K. | | Deposit date: | 2020-09-08 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of apo-FabG from Vibrio harveyi
To Be Published
|
|
5I3S
 
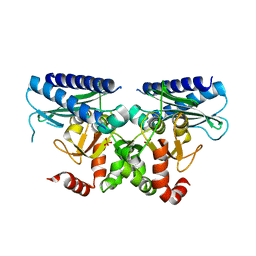 | |
6JTZ
 
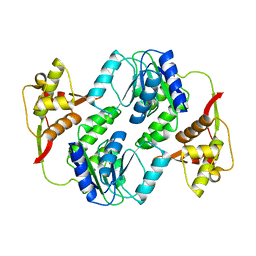 | | Crystal Structure of hRecQ1_D2-Zn-WH containing mutation on beta-hairpin | | Descriptor: | ATP-dependent DNA helicase Q1, ZINC ION | | Authors: | Das, T, Mukhopadhyay, S, Bose, M, Das, A.K, Ganguly, A. | | Deposit date: | 2019-04-12 | | Release date: | 2020-04-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.797 Å) | | Cite: | Residues at the interface between zinc binding and winged helix domains of human RECQ1 play a significant role in DNA strand annealing activity.
Nucleic Acids Res., 2021
|
|
3P7X
 
 | | Crystal structure of an atypical two-cysteine peroxiredoxin (SAOUHSC_01822) from Staphylococcus aureus NCTC8325 | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, Probable thiol peroxidase, ... | | Authors: | Bhattacharyya, S, Dutta, D, Ghosh, A.K, Das, A.K. | | Deposit date: | 2010-10-13 | | Release date: | 2011-10-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure of an atypical two-cysteine peroxiredoxin (SAOUHSC_01822) from Staphylococcus aureus NCTC8325
To be Published
|
|
6A4J
 
 | | Crystal structure of Thioredoxin reductase 2 from Staphylococcus aureus | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin--NADP reductase | | Authors: | Bose, M, Bhattacharyya, S, Ghosh, A.K, Das, A.K. | | Deposit date: | 2018-06-20 | | Release date: | 2018-07-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Elucidation of the mechanism of disulfide exchange between staphylococcal thioredoxin2 and thioredoxin reductase2: A structural insight.
Biochimie, 160, 2019
|
|
3QMF
 
 | | Crystal strucuture of an inositol monophosphatase family protein (SAS2203) from Staphylococcus aureus MSSA476 | | Descriptor: | Inositol monophosphatase family protein, SULFATE ION | | Authors: | Bhattacharyya, S, Dutta, D, Ghosh, A.K, Das, A.K. | | Deposit date: | 2011-02-04 | | Release date: | 2012-01-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Staphylococcal dual specific inositol monophosphatase/NADP(H) phosphatase (SAS2203) delineates the molecular basis of substrate specificity
Biochimie, 94, 2012
|
|
6JQY
 
 | |
