1VJI
 
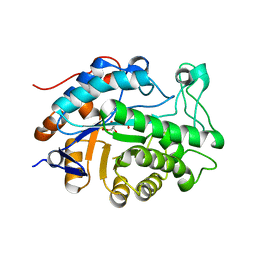 | | Gene Product of At1g76680 from Arabidopsis thaliana | | Descriptor: | 12-oxophytodienoate reductase (OPR1), FLAVIN MONONUCLEOTIDE | | Authors: | Wesenberg, G.E, Smith, D.W, Phillips Jr, G.N, Johnson, K.A, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-02-24 | | Release date: | 2004-03-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | X-ray structure of Arabidopsis At1g77680, 12-oxophytodienoate reductase isoform 1.
Proteins, 61, 2005
|
|
1YDA
 
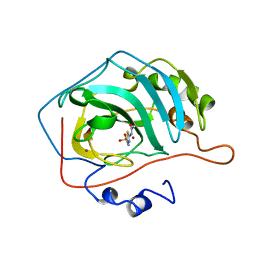 | | STRUCTURAL BASIS OF INHIBITOR AFFINITY TO VARIANTS OF HUMAN CARBONIC ANHYDRASE II | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Nair, S.K, Christianson, D.W. | | Deposit date: | 1994-12-22 | | Release date: | 1995-02-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of inhibitor affinity to variants of human carbonic anhydrase II.
Biochemistry, 34, 1995
|
|
1YHP
 
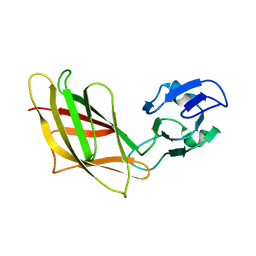 | | Solution Structure of Ca2+-free DdCAD-1 | | Descriptor: | Calcium-dependent cell adhesion molecule-1 | | Authors: | Lin, Z, Huang, H.B, Siu, C.H, Yang, D.W. | | Deposit date: | 2005-01-10 | | Release date: | 2006-01-31 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the adhesion molecule DdCAD-1 reveal new insights into Ca(2+)-dependent cell-cell adhesion
Nat.Struct.Mol.Biol., 13, 2006
|
|
1YJ4
 
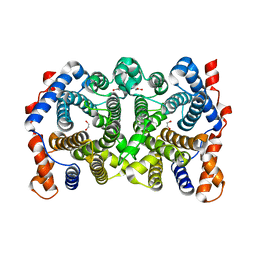 | | Y305F Trichodiene Synthase | | Descriptor: | 1,2-ETHANEDIOL, Trichodiene synthase | | Authors: | Vedula, L.S, Rynkiewicz, M.J, Pyun, H.J, Coates, R.M, Cane, D.E, Christianson, D.W. | | Deposit date: | 2005-01-13 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular Recognition of the Substrate Diphosphate Group Governs Product Diversity in Trichodiene Synthase Mutants.
Biochemistry, 44, 2005
|
|
4MHL
 
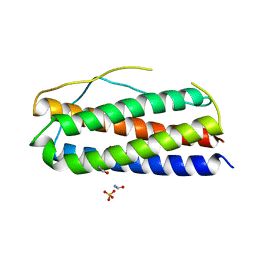 | | The crystal structure of human interleukin-11 | | Descriptor: | FORMAMIDE, Interleukin-11, SULFATE ION | | Authors: | Griffin, M.D.W. | | Deposit date: | 2013-08-29 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The structure of human interleukin-11 reveals receptor-binding site features and structural differences from interleukin-6.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2UU8
 
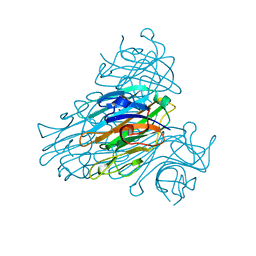 | | X-ray structure of Ni, Ca concanavalin A at Ultra-high resolution (0. 94A) | | Descriptor: | CALCIUM ION, CONCANAVALIN, NICKEL (II) ION | | Authors: | Ahmed, H.U, Blakeley, M.P, Cianci, M, Cruickshank, D.W.J, Hubbard, J.A, Helliwell, J.R. | | Deposit date: | 2007-03-01 | | Release date: | 2007-07-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | The Determination of Protonation States in Proteins.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2V3H
 
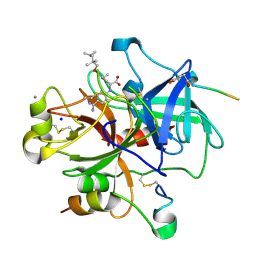 | | Thrombin with 3-cycle no F | | Descriptor: | (2R)-({4-[AMINO(IMINO)METHYL]PHENYL}AMINO){3-[3-(DIMETHYLAMINO)-2,2-DIMETHYLPROPOXY]-5-ETHYLPHENYL}ACETIC ACID, CALCIUM ION, HIRUDIN IIA, ... | | Authors: | Banner, D.W, Obst, U. | | Deposit date: | 2007-06-18 | | Release date: | 2008-06-24 | | Last modified: | 2013-10-23 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Fluorine in Pharmaceuticals: Looking Beyond Intuition.
Science, 317, 2007
|
|
2V3O
 
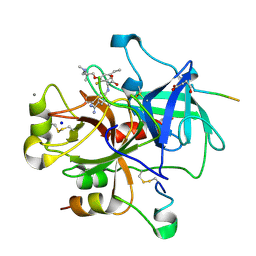 | | Thrombin with 3-cycle with F | | Descriptor: | (2R)-[(4-CARBAMIMIDOYLPHENYL)AMINO]{3-[3-(DIMETHYLAMINO)-2,2-DIMETHYLPROPOXY]-5-ETHYL-2-FLUOROPHENYL}ETHANOIC ACID, CALCIUM ION, HIRUDIN IIA, ... | | Authors: | Banner, D.W, Wessel, H.P. | | Deposit date: | 2007-06-19 | | Release date: | 2008-06-24 | | Last modified: | 2013-10-23 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Fluorine in Pharmaceuticals: Looking Beyond Intuition.
Science, 317, 2007
|
|
2UUJ
 
 | | Thrombin-hirugen-gw473178 ternary complex at 1.32A resolution | | Descriptor: | CALCIUM ION, HIRUDIN I, HUMAN ALPHA THROMBIN, ... | | Authors: | Ahmed, H.U, Blakeley, M.P, Cianci, M, Cruickshank, D.W.J, Hubbard, J.A, Helliwell, J.R. | | Deposit date: | 2007-03-03 | | Release date: | 2007-09-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | The Determination of Protonation States in Proteins.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2UUK
 
 | | Thrombin-hirugen-gw420128 ternary complex at 1.39A resolution | | Descriptor: | CALCIUM ION, HIRUDIN I, HUMAN ALPHA THROMBIN, ... | | Authors: | Ahmed, H.U, Blakeley, M.P, Cianci, M, Cruickshank, D.W.J, Hubbard, J.A, Helliwell, J.R. | | Deposit date: | 2007-03-03 | | Release date: | 2007-09-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | The Determination of Protonation States in Proteins.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2UUF
 
 | | Thrombin-hirugen binary complex at 1.26A resolution | | Descriptor: | CALCIUM ION, HIRUDIN I, HUMAN ALPHA THROMBIN, ... | | Authors: | Ahmed, H.U, Blakeley, M.P, Cianci, M, Cruickshank, D.W.J, Hubbard, J.A, Helliwell, J.R. | | Deposit date: | 2007-03-02 | | Release date: | 2007-09-04 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | The Determination of Protonation States in Proteins.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
7FF1
 
 | | Structure of C34E136G/N36 | | Descriptor: | gp41 C34E136G, gp41 N36 | | Authors: | Yu, D.W, Qin, B. | | Deposit date: | 2021-07-22 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.689 Å) | | Cite: | Structure of C34E136G/N36
To Be Published
|
|
7JHY
 
 | | Type IV-B CRISPR Complex | | Descriptor: | Csf2 (Cas7), Csf4 (Cas11), RNA (31-MER) | | Authors: | Bravo, J.P.K, Taylor, D.W. | | Deposit date: | 2020-07-21 | | Release date: | 2021-03-31 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of a type IV CRISPR-Cas ribonucleoprotein complex.
Iscience, 24, 2021
|
|
7JJN
 
 | | Eubacterium rectale Amy13B (EUR_01860) | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Koropatkin, N.M, Cockburn, D.W, Cerqueira, F.M. | | Deposit date: | 2020-07-27 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The structures of the GH13_36 amylases from Eubacterium rectale and Ruminococcus bromii reveal subsite architectures that favor maltose production
Amylase, 4, 2020
|
|
1SSW
 
 | | Crystal structure of phage T4 lysozyme mutant Y24A/Y25A/T26A/I27A/C54T/C97A | | Descriptor: | BETA-MERCAPTOETHANOL, Lysozyme | | Authors: | He, M.M, Baase, W.A, Xiao, H, Heinz, D.W, Matthews, B.W. | | Deposit date: | 2004-03-24 | | Release date: | 2004-10-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Alanine-scanning mutagenesis of the beta-sheet region of phage T4 lysozyme suggests that tertiary context has a dominant effect on beta-sheet formation
Protein Sci., 13, 2004
|
|
1SSY
 
 | | Crystal structure of phage T4 lysozyme mutant G28A/I29A/G30A/C54T/C97A | | Descriptor: | Lysozyme | | Authors: | He, M.M, Baase, W.A, Xiao, H, Heinz, D.W, Matthews, B.W. | | Deposit date: | 2004-03-24 | | Release date: | 2004-10-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Alanine-scanning mutagenesis of the beta-sheet region of phage T4 lysozyme suggests that tertiary context has a dominant effect on beta-sheet formation
Protein Sci., 13, 2004
|
|
1TDT
 
 | |
1HQ4
 
 | | STRUCTURE OF NATIVE CATALYTIC ANTIBODY HA5-19A4 | | Descriptor: | ANTIBODY HA5-19A4 FAB HEAVY CHAIN, ANTIBODY HA5-19A4 FAB LIGHT CHAIN, CADMIUM ION | | Authors: | Paschall, C.M, Hasserodt, J, Lerner, R.A, Janda, K.D, Christianson, D.W. | | Deposit date: | 2000-12-14 | | Release date: | 2003-06-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Polyene Cyclization Reactions
Thesis, 2000
|
|
1ZWJ
 
 | | X-ray structure of galt-like protein from arabidopsis thaliana AT5G18200 | | Descriptor: | ZINC ION, putative galactose-1-phosphate uridyl transferase | | Authors: | Wesenberg, G.E, Smith, D.W, Phillips Jr, G.N, McCoy, J.G, Johnson, K.A, Bitto, E, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-06-03 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Mechanism of an ADP-Glucose Phosphorylase from
Arabidopsis thaliana
Biochemistry, 45, 2006
|
|
3R4D
 
 | | Crystal structure of mouse coronavirus receptor-binding domain complexed with its murine receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CEA-related cell adhesion molecule 1, ... | | Authors: | Peng, G.Q, Sun, D.W, Rajashankar, K.R, Qian, Z.H, Holmes, K.V, Li, F. | | Deposit date: | 2011-03-17 | | Release date: | 2011-06-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of mouse coronavirus receptor-binding domain complexed with its murine receptor.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3S1A
 
 | | Crystal structure of the phosphorylation-site double mutant S431E/T432E of the KaiC circadian clock protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein kinase kaiC, MAGNESIUM ION | | Authors: | Pattanayek, R, Williams, D.W, Rossi, G, Weigand, S, Mori, T, Johnson, C.H, Stewart, P.L, Egli, M. | | Deposit date: | 2011-05-14 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Combined SAXS/EM Based Models of the S. elongatus Post-Translational Circadian Oscillator and its Interactions with the Output His-Kinase SasA.
Plos One, 6, 2011
|
|
7EHS
 
 | | Levansucrase from Brenneria sp. EniD 312 | | Descriptor: | GLYCEROL, Levansucrase, NONAETHYLENE GLYCOL, ... | | Authors: | Xu, W, Ni, D.W, Hou, X.D, Rao, Y.J, Pijning, T, Guskov, A, Mu, W.M. | | Deposit date: | 2021-03-30 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Levansucrase from the Gram-Negative Bacterium Brenneria Provides Insights into Its Product Size Specificity.
J.Agric.Food Chem., 70, 2022
|
|
7EHR
 
 | | Levansucrase from Brenneria sp. EniD 312 at 1.33 angstroms resolution | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Xu, W, Ni, D.W, Hou, X.D, Rao, Y.J, Pijning, T, Guskov, A, Mu, W.M. | | Deposit date: | 2021-03-30 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Crystal Structure of Levansucrase from the Gram-Negative Bacterium Brenneria Provides Insights into Its Product Size Specificity.
J.Agric.Food Chem., 70, 2022
|
|
7F0R
 
 | | Cryo-EM structure of Pseudomonas aeruginosa SutA transcription activation complex | | Descriptor: | DNA (70-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | He, D.W, You, L.L, Zhang, Y. | | Deposit date: | 2021-06-06 | | Release date: | 2022-07-27 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Pseudomonas aeruginosa SutA wedges RNAP lobe domain open to facilitate promoter DNA unwinding.
Nat Commun, 13, 2022
|
|
