4FDX
 
 | |
6B3P
 
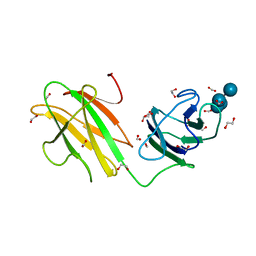 | | Crystal structure of CBMbc (family CBM26) from Eubacterium rectale Amy13K in Complex with Maltoheptaose | | 分子名称: | 1,2-ETHANEDIOL, Amy13K, FORMIC ACID, ... | | 著者 | Cockburn, D.W, Wawrzak, Z, Perez Medina, K, Koropatkin, N.M. | | 登録日 | 2017-09-22 | | 公開日 | 2017-11-29 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Novel carbohydrate binding modules in the surface anchored alpha-amylase of Eubacterium rectale provide a molecular rationale for the range of starches used by this organism in the human gut.
Mol. Microbiol., 107, 2018
|
|
7U8Z
 
 | | Crystal structure of Danio rerio histone deacetylase 6 catalytic domain 2 (CD2) complexed with fluorinated peptoid inhibitor | | 分子名称: | 1,2-ETHANEDIOL, 4-({N-[2-(benzylamino)-2-oxoethyl]-4-(dimethylamino)benzamido}methyl)-3-fluoro-N-hydroxybenzamide, ACETATE ION, ... | | 著者 | Watson, P.R, Cragin, A.D, Christianson, D.W. | | 登録日 | 2022-03-09 | | 公開日 | 2022-11-23 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Development of Fluorinated Peptoid-Based Histone Deacetylase (HDAC) Inhibitors for Therapy-Resistant Acute Leukemia.
J.Med.Chem., 65, 2022
|
|
4FAZ
 
 | |
4RLA
 
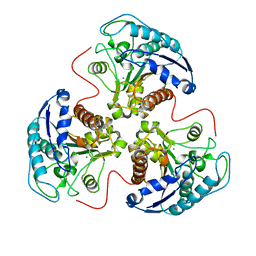 | |
3HPQ
 
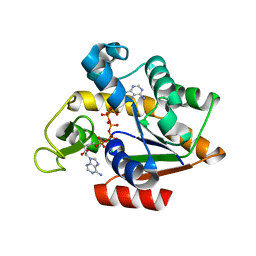 | |
7U59
 
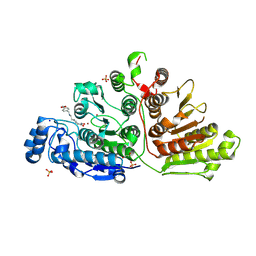 | |
6C6K
 
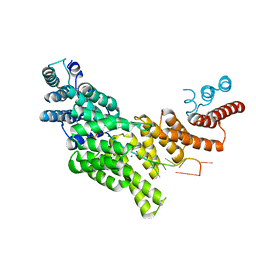 | | Structural basis for preferential recognition of cap 0 RNA by a human IFIT1-IFIT3 protein complex | | 分子名称: | Interferon-induced protein with tetratricopeptide repeats 1, Interferon-induced protein with tetratricopeptide repeats 3, MAGNESIUM ION, ... | | 著者 | Amarasinghe, G.K, Leung, D.W, Johnson, B, Xu, W. | | 登録日 | 2018-01-18 | | 公開日 | 2018-04-04 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.54 Å) | | 主引用文献 | Human IFIT3 Modulates IFIT1 RNA Binding Specificity and Protein Stability.
Immunity, 48, 2018
|
|
4FBY
 
 | | fs X-ray diffraction of Photosystem II | | 分子名称: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | 著者 | Kern, J, Alonso-Mori, R, Hellmich, J, Tran, R, Hattne, J, Laksmono, H, Gloeckner, C, Echols, N, Sierra, R.G, Sellberg, J, Lassalle-Kaiser, B, Gildea, R.J, Glatzel, P, Grosse-Kunstleve, R.W, Latimer, M.J, Mcqueen, T.A, Difiore, D, Fry, A.R, Messerschmidt, M.M, Miahnahri, A, Schafer, D.W, Seibert, M.M, Sokaras, D, Weng, T.-C, Zwart, P.H, White, W.E, Adams, P.D, Bogan, M.J, Boutet, S, Williams, G.J, Messinger, J, Sauter, N.K, Zouni, A, Bergmann, U, Yano, J, Yachandra, V.K. | | 登録日 | 2012-05-23 | | 公開日 | 2012-06-20 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (6.56 Å) | | 主引用文献 | Room temperature femtosecond X-ray diffraction of photosystem II microcrystals.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4FCI
 
 | |
4U0H
 
 | | Crystal Structure of M. tuberculosis ClpP1P1 | | 分子名称: | ATP-dependent Clp protease proteolytic subunit 1, SULFATE ION | | 著者 | Schmitz, K.R, Carney, D.W, Sello, J.K, Sauer, R.T. | | 登録日 | 2014-07-11 | | 公開日 | 2014-10-08 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (3.2479 Å) | | 主引用文献 | Crystal structure of Mycobacterium tuberculosis ClpP1P2 suggests a model for peptidase activation by AAA+ partner binding and substrate delivery.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4TZA
 
 | |
4EP9
 
 | | CRYSTAL STRUCTURE OF RAT CARNITINE PALMITOYLTRANSFERASE 2 IN COMPLEX WITH CoA-site inhibitor | | 分子名称: | 4-[({1-[(5-chloro-2-methoxyphenyl)sulfonyl]-4-methyl-2,3-dihydro-1H-indol-6-yl}carbonyl)amino]benzoic acid, Carnitine O-palmitoyltransferase 2, mitochondrial, ... | | 著者 | Rufer, A.C, Thoma, R, Benz, J, Stihle, M, Gsell, B, De Roo, E, Banner, D.W, Mueller, F, Chomienne, O, Hennig, M. | | 登録日 | 2012-04-17 | | 公開日 | 2013-04-17 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Isothermal titration calorimetry with micelles: Thermodynamics of inhibitor binding to carnitine palmitoyltransferase 2 membrane protein.
FEBS Open Bio, 3, 2013
|
|
4TZG
 
 | | Crystal structure of eCGP123, an extremely thermostable green fluorescent protein | | 分子名称: | Fluorescent Protein | | 著者 | Close, D.W, Don Paul, C, Traore, D.A.K, Wilce, M.C.J, Prescott, M, Bradbury, A.R.M. | | 登録日 | 2014-07-10 | | 公開日 | 2014-10-22 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Thermal green protein, an extremely stable, nonaggregating fluorescent protein created by structure-guided surface engineering.
Proteins, 83, 2015
|
|
3HPR
 
 | |
4FCK
 
 | |
4UIL
 
 | | crystal structure of quinine-dependent Fab 314.1 with quinine | | 分子名称: | FAB 314.1, Quinine | | 著者 | Zhu, J, Zhu, J, Bougie, D.W, Aster, R.H, Springer, T.A. | | 登録日 | 2015-03-30 | | 公開日 | 2015-09-02 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.853 Å) | | 主引用文献 | Structural Basis for Quinine-Dependent Antibody Binding to Platelet Integrin Alphaiib Beta3
Blood, 126, 2015
|
|
3BNY
 
 | | Crystal structure of aristolochene synthase complexed with 2-fluorofarnesyl diphosphate (2F-FPP) | | 分子名称: | (2Z,6E)-2-fluoro-3,7,11-trimethyldodeca-2,6,10-trien-1-yl trihydrogen diphosphate, Aristolochene synthase, BETA-MERCAPTOETHANOL, ... | | 著者 | Shishova, E.Y, Christianson, D.W. | | 登録日 | 2007-12-14 | | 公開日 | 2008-03-25 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | X-ray Crystallographic Studies of Substrate Binding to Aristolochene Synthase Suggest a Metal Ion Binding Sequence for Catalysis
J.Biol.Chem., 283, 2008
|
|
4UIM
 
 | | crystal structure of quinine-dependent Fab 314.3 | | 分子名称: | FAB 314.3, SULFATE ION | | 著者 | Zhu, J, Zhu, J, Bougie, D.W, Aster, R.H, Springer, T.A. | | 登録日 | 2015-03-30 | | 公開日 | 2015-09-02 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural Basis for Quinine-Dependent Antibody Binding to Platelet Integrin Alphaiib Beta3
Blood, 126, 2015
|
|
3COQ
 
 | | Structural Basis for Dimerization in DNA Recognition by Gal4 | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*DAP*DCP*DCP*DGP*DGP*DAP*DGP*DGP*DAP*DCP*DAP*DGP*DTP*DCP*DCP*DTP*DCP*DCP*DGP*DG)-3'), DNA (5'-D(*DTP*DCP*DCP*DGP*DGP*DAP*DGP*DGP*DAP*DCP*DTP*DGP*DTP*DCP*DCP*DTP*DCP*DCP*DGP*DG)-3'), ... | | 著者 | Hong, M, Fitzgerald, M.X, Harper, S, Luo, C, Speicher, D.W. | | 登録日 | 2008-03-29 | | 公開日 | 2008-07-01 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural basis for dimerization in DNA recognition by gal4.
Structure, 16, 2008
|
|
4UIK
 
 | | crystal structure of quinine-dependent Fab 314.1 | | 分子名称: | FAB 314.1 | | 著者 | Zhu, J, Zhu, J, Bougie, D.W, Aster, R.H, Springer, T.A. | | 登録日 | 2015-03-30 | | 公開日 | 2015-09-02 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural Basis for Quinine-Dependent Antibody Binding to Platelet Integrin Alphaiib Beta3
Blood, 126, 2015
|
|
4U0G
 
 | | Crystal Structure of M. tuberculosis ClpP1P2 bound to ADEP and agonist | | 分子名称: | ADEP-2B5Me, ATP-dependent Clp protease proteolytic subunit 1, ATP-dependent Clp protease proteolytic subunit 2, ... | | 著者 | Schmitz, K.R, Carney, D.W, Sello, J.K, Sauer, R.T. | | 登録日 | 2014-07-11 | | 公開日 | 2014-10-08 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (3.1978 Å) | | 主引用文献 | Crystal structure of Mycobacterium tuberculosis ClpP1P2 suggests a model for peptidase activation by AAA+ partner binding and substrate delivery.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3COT
 
 | | Crystal structure of human liver delta(4)-3-ketosteroid 5beta-reductase (akr1d1) in complex with progesterone and nadp. Resolution: 2.03 A. | | 分子名称: | 3-oxo-5-beta-steroid 4-dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROGESTERONE | | 著者 | Di Costanzo, L, Drury, J, Penning, T.M, Christianson, D.W. | | 登録日 | 2008-03-29 | | 公開日 | 2008-04-08 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Crystal Structure of Human Liver {Delta}4-3-Ketosteroid 5{beta}-Reductase (AKR1D1) and Implications for Substrate Binding and Catalysis.
J.Biol.Chem., 283, 2008
|
|
6CL1
 
 | |
4UIN
 
 | | crystal structure of quinine-dependent Fab 314.3 with quinine | | 分子名称: | FAB 314.3, Quinine | | 著者 | Zhu, J, Zhu, J, Bougie, D.W, Aster, R.H, Springer, T.A. | | 登録日 | 2015-03-30 | | 公開日 | 2015-09-02 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural Basis for Quinine-Dependent Antibody Binding to Platelet Integrin Alphaiib Beta3
Blood, 126, 2015
|
|
