5LQO
 
 | |
5LR4
 
 | |
5O62
 
 | |
5O69
 
 | | The structure of the thermobifida fusca guanidine III riboswitch with agmatine. | | Descriptor: | AGMATINE, MAGNESIUM ION, RNA (37-MER), ... | | Authors: | Huang, L, Wang, J, Lilley, D.M.J. | | Deposit date: | 2017-06-06 | | Release date: | 2017-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.319 Å) | | Cite: | Structure of the Guanidine III Riboswitch.
Cell Chem Biol, 24, 2017
|
|
2YDS
 
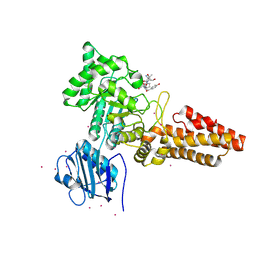 | | CpOGA D298N in complex with TAB1-derived O-GlcNAc peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, O-GLCNACASE NAGJ, ... | | Authors: | Schimpl, M, Borodkin, V.S, Gray, L.J, van Aalten, D.M.F. | | Deposit date: | 2011-03-24 | | Release date: | 2012-03-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Synergy of Peptide and Sugar in O-Glcnacase Substrate Recognition.
Chem.Biol., 19, 2012
|
|
5VJ7
 
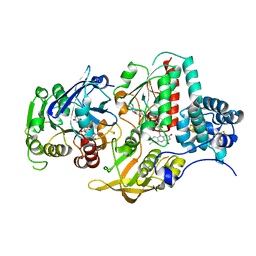 | | Ferredoxin NADP Oxidoreductase (Xfn) | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin-NADP(+) reductase subunit alpha, ... | | Authors: | Zadvornyy, O.A, Nguyen, D.M.N, Schut, G.J, Lipscomb, G.L, Tokmina-Lukaszewska, M, Adams, M.W.W, Peters, J.W. | | Deposit date: | 2017-04-18 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Two functionally distinct NADP(+)-dependent ferredoxin oxidoreductases maintain the primary redox balance of Pyrococcus furiosus.
J. Biol. Chem., 292, 2017
|
|
5T6F
 
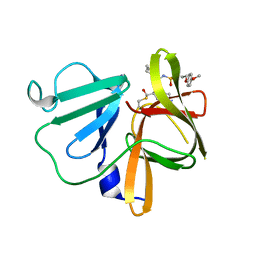 | | 1.90 A resolution structure of Norovirus 3CL protease in complex with the dipeptidyl inhibitor 7l (orthorhombic P form) | | Descriptor: | 3-cyclohexyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-N~2~-{[3-(4-methoxyphenoxy)propyl]sulfonyl}-L- alaninamide, Genome polyprotein | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Kankanamalage, A.C.G, Kim, Y, Rathnayake, A.D, Damalanka, V.C, Weerawarna, P.M, Doyle, S.T, Alsoudi, A.F, Dissanayake, D.M.P, Chang, K.-O, Groutas, W.C. | | Deposit date: | 2016-09-01 | | Release date: | 2016-11-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based exploration and exploitation of the S4 subsite of norovirus 3CL protease in the design of potent and permeable inhibitors.
Eur J Med Chem, 126, 2016
|
|
5T6D
 
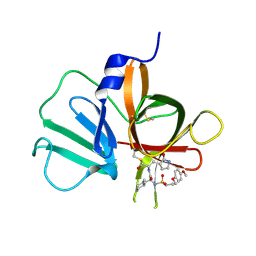 | | 2.10 A resolution structure of Norovirus 3CL protease in complex with the dipeptidyl inhibitor 7l (hexagonal form) | | Descriptor: | 3-cyclohexyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-N~2~-{[3-(4-methoxyphenoxy)propyl]sulfonyl}-L- alaninamide, Genome polyprotein | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Kankanamalage, A.C.G, Kim, Y, Rathnayake, A.D, Damalanka, V.C, Weerawarna, P.M, Doyle, S.T, Alsoudi, A.F, Dissanayake, D.M.P, Chang, K.-O, Groutas, W.C. | | Deposit date: | 2016-09-01 | | Release date: | 2016-11-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based exploration and exploitation of the S4 subsite of norovirus 3CL protease in the design of potent and permeable inhibitors.
Eur J Med Chem, 126, 2016
|
|
5T6G
 
 | | 2.45 A resolution structure of Norovirus 3CL protease in complex with the dipeptidyl inhibitor 7m (hexagonal form) | | Descriptor: | 3-cyclohexyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-N~2~-(octylsulfonyl)-L-alaninamide, Genome polyprotein | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Kankanamalage, A.C.G, Kim, Y, Rathnayake, A.D, Damalanka, V.C, Weerawarna, P.M, Doyle, S.T, Alsoudi, A.F, Dissanayake, D.M.P, Chang, K.-O, Groutas, W.C. | | Deposit date: | 2016-09-01 | | Release date: | 2016-11-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure-based exploration and exploitation of the S4 subsite of norovirus 3CL protease in the design of potent and permeable inhibitors.
Eur J Med Chem, 126, 2016
|
|
1HJW
 
 | | Crystal structure of hcgp-39 in complex with chitin octamer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Houston, D.R, Recklies, A.D, Krupa, J.C, Van Aalten, D.M.F. | | Deposit date: | 2003-02-28 | | Release date: | 2003-03-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Ligand-Induced Conformational Change of the 39-kDa Glycoprotein from Human Articular Chondrocytes
J.Biol.Chem., 278, 2003
|
|
1IKT
 
 | |
1JPD
 
 | | L-Ala-D/L-Glu Epimerase | | Descriptor: | L-Ala-D/L-Glu Epimerase | | Authors: | Gulick, A.M, Schmidt, D.M.Z, Gerlt, J.A, Rayment, I. | | Deposit date: | 2001-08-01 | | Release date: | 2001-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: crystal structures of the L-Ala-D/L-Glu epimerases from Escherichia coli and Bacillus subtilis.
Biochemistry, 40, 2001
|
|
1JPM
 
 | | L-Ala-D/L-Glu Epimerase | | Descriptor: | GLYCEROL, L-Ala-D/L-Glu Epimerase, MAGNESIUM ION | | Authors: | Gulick, A.M, Schmidt, D.M.Z, Gerlt, J.A, Rayment, I. | | Deposit date: | 2001-08-02 | | Release date: | 2001-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: crystal structures of the L-Ala-D/L-Glu epimerases from Escherichia coli and Bacillus subtilis.
Biochemistry, 40, 2001
|
|
1KOU
 
 | | Crystal Structure of the Photoactive Yellow Protein Reconstituted with Caffeic Acid at 1.16 A Resolution | | Descriptor: | CAFFEIC ACID, N-BUTANE, PHOTOACTIVE YELLOW PROTEIN | | Authors: | van Aalten, D.M.F, Crielaard, W, Hellingwerf, K.J, Joshua-Tor, L. | | Deposit date: | 2001-12-22 | | Release date: | 2002-04-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Structure of the photoactive yellow protein reconstituted with caffeic acid at 1.16 A resolution.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1KWF
 
 | | Atomic Resolution Structure of an Inverting Glycosidase in Complex with Substrate | | Descriptor: | Endoglucanase A, beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Guerin, D.M.A, Lascombe, M.-B, Costabel, M, Souchon, H, Lamzin, V, Beguin, P, Alzari, P.M. | | Deposit date: | 2002-01-29 | | Release date: | 2002-03-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Atomic (0.94 A) resolution structure of an inverting glycosidase in complex with substrate.
J.Mol.Biol., 316, 2002
|
|
2EU1
 
 | | Crystal structure of the chaperonin GroEL-E461K | | Descriptor: | GROEL | | Authors: | Cabo-Bilbao, A, Spinelli, S, Sot, B, Agirre, J, Mechaly, A.E, Muga, A, Guerin, D.M.A. | | Deposit date: | 2005-10-28 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Crystal structure of the temperature-sensitive and allosteric-defective chaperonin GroEL(E461K).
J.Struct.Biol., 155, 2006
|
|
2H6E
 
 | | Crystal structure of the D-arabinose dehydrogenase from Sulfolobus solfataricus | | Descriptor: | D-arabinose 1-dehydrogenase, ZINC ION | | Authors: | Brouns, S.J.J, Turnbull, A.P, Akerboom, J, Willemen, H.L.D.M, De Vos, W.M, Van der Oost, J. | | Deposit date: | 2006-05-31 | | Release date: | 2007-06-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure and Biochemical Properties of the d-Arabinose Dehydrogenase from Sulfolobus solfataricus
J.Mol.Biol., 371, 2007
|
|
3CHE
 
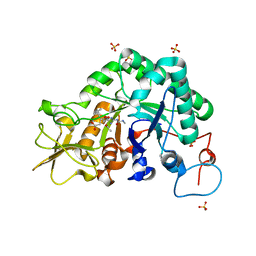 | |
3CHF
 
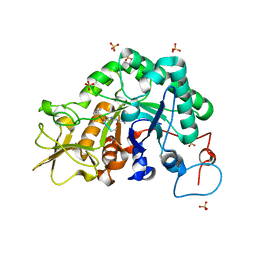 | |
3CHC
 
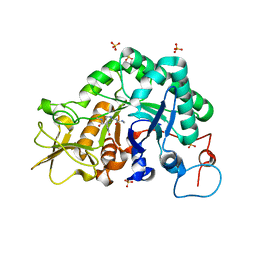 | |
3CH9
 
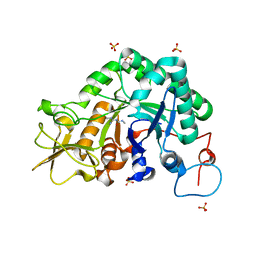 | |
2J62
 
 | | Structure of a bacterial O-glcnacase in complex with glcnacstatin | | Descriptor: | CHLORIDE ION, N-[(5R,6R,7R,8S)-6,7-DIHYDROXY-5-(HYDROXYMETHYL)-2-(2-PHENYLETHYL)-1,5,6,7,8,8A-HEXAHYDROIMIDAZO[1,2-A]PYRIDIN-8-YL]-2-METHYLPROPANAMIDE, O-GlcNAcase NagJ | | Authors: | Dorfmueller, H.C, Borodkin, V.S, Schimpl, M, Shepherd, S.M, Shpiro, N.A, van Aalten, D.M.F. | | Deposit date: | 2006-09-22 | | Release date: | 2007-02-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | GlcNAcstatin: a picomolar, selective O-GlcNAcase inhibitor that modulates intracellular O-glcNAcylation levels.
J. Am. Chem. Soc., 128, 2006
|
|
2IUZ
 
 | | Crystal structure of Aspergillus fumigatus chitinase B1 in complex with C2-dicaffeine | | Descriptor: | 1,1'-ETHANE-1,2-DIYLBIS(3,7-DIMETHYL-3,7-DIHYDRO-1H-PURINE-2,6-DIONE), CHITINASE, SULFATE ION | | Authors: | Schuttelkopf, A.W, Andersen, O.A, Rao, F.V, Allwood, M, Lloyd, C.M, Eggleston, I.M, Van Aalten, D.M.F. | | Deposit date: | 2006-06-08 | | Release date: | 2006-06-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Screening-Based Discovery and Structural Dissection of a Novel Family 18 Chitinase Inhibitor
J.Biol.Chem., 281, 2006
|
|
2IW0
 
 | | Structure of the chitin deacetylase from the fungal pathogen Colletotrichum lindemuthianum | | Descriptor: | ACETATE ION, CHITIN DEACETYLASE, CHLORIDE ION, ... | | Authors: | Blair, D.E, Hekmat, O, Schuttelkopf, A.W, Shrestha, B, Tokuyasu, K, Withers, S.G, van Aalten, D.M.F. | | Deposit date: | 2006-06-23 | | Release date: | 2006-07-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure and Mechanism of Chitin Deacetylase from the Fungal Pathogen Colletotrichum Lindemuthianum.
Biochemistry, 45, 2006
|
|
3CHD
 
 | |
