8ORQ
 
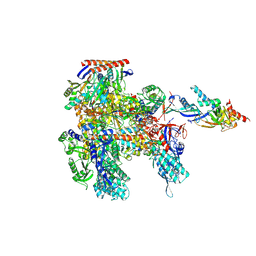 | | Cryo-EM structure of Pyrococcus furiosus apo form RNA polymerase open clamp conformation | | Descriptor: | DNA-directed RNA polymerase subunit Rpo10, DNA-directed RNA polymerase subunit Rpo11, DNA-directed RNA polymerase subunit Rpo12, ... | | Authors: | Tarau, D.M, Reichelt, R, Heiss, F.B, Pilsl, M, Hausner, W, Engel, C, Grohmann, D. | | Deposit date: | 2023-04-17 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of archaeal RNA polymerase transcription elongation and Spt4/5 recruitment.
Nucleic Acids Res., 52, 2024
|
|
8P2I
 
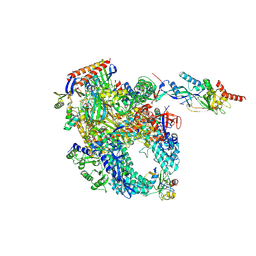 | | Cryo-EM structure of Pyrococcus furiosus apo form RNA polymerase contracted clamp conformation with Spt4/5 | | Descriptor: | DNA-directed RNA polymerase subunit Rpo10, DNA-directed RNA polymerase subunit Rpo11, DNA-directed RNA polymerase subunit Rpo12, ... | | Authors: | Tarau, D.M, Reichelt, R, Heiss, F.B, Pilsl, M, Hausner, W, Engel, C, Grohmann, D. | | Deposit date: | 2023-05-16 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of archaeal RNA polymerase transcription elongation and Spt4/5 recruitment.
Nucleic Acids Res., 52, 2024
|
|
8OKI
 
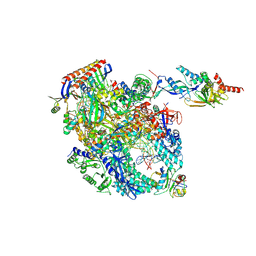 | | Cryo-EM structure of Pyrococcus furiosus transcription elongation complex bound to Spt4/5 | | Descriptor: | DNA Non-Template Strand, DNA Template Strand, DNA-directed RNA polymerase subunit Rpo10, ... | | Authors: | Tarau, D.M, Reichelt, R, Heiss, F.B, Pilsl, M, Hausner, W, Engel, C, Grohmann, D. | | Deposit date: | 2023-03-28 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structural basis of archaeal RNA polymerase transcription elongation and Spt4/5 recruitment.
Nucleic Acids Res., 52, 2024
|
|
8RBO
 
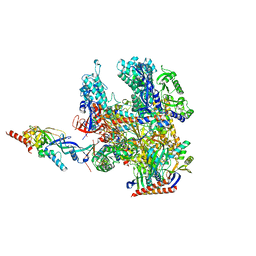 | | Cryo-EM structure of Pyrococcus furiosus apo form RNA polymerase contracted clamp conformation | | Descriptor: | DNA-directed RNA polymerase subunit Rpo10, DNA-directed RNA polymerase subunit Rpo11, DNA-directed RNA polymerase subunit Rpo12, ... | | Authors: | Tarau, D.M, Reichelt, R, Heiss, F.B, Pilsl, M, Hausner, W, Engel, C, Grohmann, D. | | Deposit date: | 2023-12-04 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structural basis of archaeal RNA polymerase transcription elongation and Spt4/5 recruitment.
Nucleic Acids Res., 52, 2024
|
|
8H8Y
 
 | | Crystal structure of AbHheG from Acidimicrobiia bacterium | | Descriptor: | GLYCEROL, alpha/beta hydrolase | | Authors: | Zhou, C.H, Chen, X, Han, X, Liu, W.D, Wu, Q.Q, Zhu, D.M, Ma, Y.H. | | Deposit date: | 2022-10-24 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Flipping the Substrate Creates a Highly Selective Halohydrin Dehalogenase for the Synthesis of Chiral 4-Aryl-2-oxazolidinones from Readily Available Epoxides
Acs Catalysis, 13, 2023
|
|
8HQP
 
 | | Crystal structure of AbHheG mutant from Acidimicrobiia bacterium | | Descriptor: | AbHheG_m | | Authors: | Zhou, C.H, Chen, X, Han, X, Liu, W.D, Wu, Q.Q, Zhu, D.M, Ma, Y.H. | | Deposit date: | 2022-12-13 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Flipping the Substrate Creates a Highly Selective Halohydrin Dehalogenase for the Synthesis of Chiral 4-Aryl-2-oxazolidinones from Readily Available Epoxides
Acs Catalysis, 13, 2023
|
|
7KJR
 
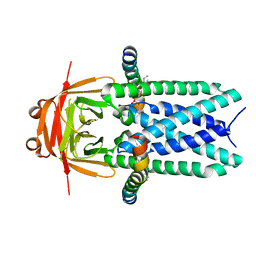 | | Cryo-EM structure of SARS-CoV-2 ORF3a | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Apolipoprotein A-I, ORF3a protein | | Authors: | Kern, D.M, Hoel, C.M, Kotecha, A, Brohawn, S.G. | | Deposit date: | 2020-10-26 | | Release date: | 2020-11-18 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.08 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 ORF3a in lipid nanodiscs.
Nat.Struct.Mol.Biol., 28, 2021
|
|
1TYC
 
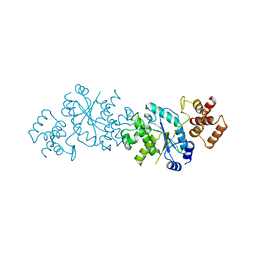 | |
1TYA
 
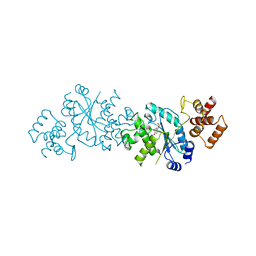 | |
1TYB
 
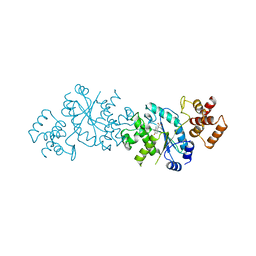 | |
8JQJ
 
 | | Crystal structure of carbonyl reductase SSCR mutant 1 from Sporobolomyces Salmonicolor | | Descriptor: | Aldehyde reductase 2 | | Authors: | Zhang, H.L, Li, Q, Liu, W.D, Chen, X, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2023-06-14 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Engineering a Carbonyl Reductase to Simultaneously Increase Activity Toward Bulky Ketone and Isopropanol for Dynamic Kinetic Asymmetric Reduction via Enzymatic Hydrogen Transfer
Acs Catalysis, 13, 2023
|
|
8JQK
 
 | | Crystal structure of a carbonyl reductase SSCR mutant from Sporobolomyces Salmonicolor | | Descriptor: | Aldehyde reductase 2 | | Authors: | Zhang, H.L, Li, Q, Liu, W.D, Chen, X, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2023-06-14 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Engineering a Carbonyl Reductase to Simultaneously Increase Activity Toward Bulky Ketone and Isopropanol for Dynamic Kinetic Asymmetric Reduction via Enzymatic Hydrogen Transfer
Acs Catalysis, 13, 2023
|
|
1XMZ
 
 | | Crystal structure of the dark state of kindling fluorescent protein kfp from anemonia sulcata | | Descriptor: | BETA-MERCAPTOETHANOL, GFP-like non-fluorescent chromoprotein FP595 chain 1, GFP-like non-fluorescent chromoprotein FP595 chain 2 | | Authors: | Quillin, M.L, Anstrom, D.M, Shu, X, O'Leary, S, Kallio, K, Chudakov, D.M, Remington, S.J. | | Deposit date: | 2004-10-04 | | Release date: | 2005-04-19 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Kindling Fluorescent Protein from Anemonia sulcata: Dark-State Structure at 1.38 Resolution
Biochemistry, 44, 2005
|
|
1TYD
 
 | |
2TS1
 
 | |
1MS6
 
 | | Dipeptide Nitrile Inhibitor Bound to Cathepsin S. | | Descriptor: | Cathepsin S, MORPHOLINE-4-CARBOXYLIC ACID [1S-(2-BENZYLOXY-1R-CYANO-ETHYLCARBAMOYL)-3-METHYL-BUTYL]AMIDE | | Authors: | Ward, Y.D, Thomson, D.S, Frye, L.L, Cywin, C.L, Morwick, T, Emmanuel, M.J, Zindell, R, McNeil, D, Bekkali, Y, Giradot, M, Hrapchak, M, DeTuri, M, Crane, K, White, D, Pav, S, Wang, Y, Hao, M.H, Grygon, C.A, Labadia, M.E, Freeman, D.M, Davidson, W, Hopkins, J.L, Brown, M.L, Spero, D.M. | | Deposit date: | 2002-09-19 | | Release date: | 2003-04-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design and synthesis of dipeptide nitriles as reversible and potent Cathepsin S inhibitors
J.Med.Chem., 45, 2002
|
|
4CHA
 
 | |
5DO7
 
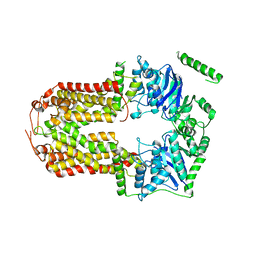 | | Crystal Structure of the Human Sterol Transporter ABCG5/ABCG8 | | Descriptor: | ATP-binding cassette sub-family G member 5, ATP-binding cassette sub-family G member 8 | | Authors: | Lee, J.-Y, Kinch, L.N, Borek, D.M, Urbatsch, I.L, Xie, X.-S, Grishin, N.V, Cohen, J.C, Otwinowski, Z, Hobbs, H.H, Rosenbaum, D.M. | | Deposit date: | 2015-09-10 | | Release date: | 2016-05-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.93 Å) | | Cite: | Crystal structure of the human sterol transporter ABCG5/ABCG8.
Nature, 533, 2016
|
|
7W3Y
 
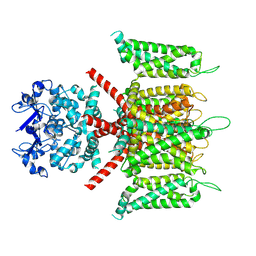 | | CryoEM structure of human Kv4.3 | | Descriptor: | Isoform 2 of Potassium voltage-gated channel subfamily D member 3 | | Authors: | Ma, D.M, Guo, J.T. | | Deposit date: | 2021-11-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for the gating modulation of Kv4.3 by auxiliary subunits.
Cell Res., 32, 2022
|
|
7W6S
 
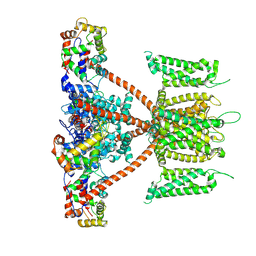 | | CryoEM structure of human KChIP2-Kv4.3 complex | | Descriptor: | Isoform 2 of Potassium voltage-gated channel subfamily D member 3, Kv channel-interacting protein 2 | | Authors: | Ma, D.M, Guo, J.T. | | Deposit date: | 2021-12-02 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for the gating modulation of Kv4.3 by auxiliary subunits.
Cell Res., 32, 2022
|
|
7W6T
 
 | | CryoEM structure of human KChIP1-Kv4.3-DPP6 complex | | Descriptor: | Dipeptidyl aminopeptidase-like protein 6, Isoform 2 of Potassium voltage-gated channel subfamily D member 3, Kv channel-interacting protein 1 | | Authors: | Ma, D.M, Guo, J.T. | | Deposit date: | 2021-12-02 | | Release date: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Structural basis for the gating modulation of Kv4.3 by auxiliary subunits.
Cell Res., 32, 2022
|
|
7W6N
 
 | | CryoEM structure of human KChIP1-Kv4.3 complex | | Descriptor: | Isoform 2 of Potassium voltage-gated channel subfamily D member 3, Kv channel-interacting protein 1 | | Authors: | Ma, D.M, Guo, J.T. | | Deposit date: | 2021-12-02 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for the gating modulation of Kv4.3 by auxiliary subunits.
Cell Res., 32, 2022
|
|
7WD3
 
 | | Cryo-EM structure of Drg1 hexamer treated with ATP and benzo-diazaborine | | Descriptor: | 2-(TOLUENE-4-SULFONYL)-2H-BENZO[D][1,2,3]DIAZABORININ-1-OL, ADENOSINE-5'-TRIPHOSPHATE, ATPase family gene 2 protein | | Authors: | Ma, C.Y, Wu, D.M, Chen, Q, Gao, N. | | Deposit date: | 2021-12-21 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural dynamics of AAA + ATPase Drg1 and mechanism of benzo-diazaborine inhibition.
Nat Commun, 13, 2022
|
|
7WBB
 
 | | Cryo-EM structure of substrate engaged Drg1 hexamer | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, AFG2 isoform 1, substrate | | Authors: | Ma, C.Y, Wu, D.M, Chen, Q, Gao, N. | | Deposit date: | 2021-12-16 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural dynamics of AAA + ATPase Drg1 and mechanism of benzo-diazaborine inhibition.
Nat Commun, 13, 2022
|
|
8CRO
 
 | | Cryo-EM structure of Pyrococcus furiosus transcription elongation complex | | Descriptor: | DNA Non-Template Strand, DNA Template Strand, DNA-directed RNA polymerase subunit Rpo10, ... | | Authors: | Tarau, D.M, Grunberger, F, Reichelt, R, Heiss, F.B, Pilsl, M, Hausner, W, Engel, C, Grohmann, D. | | Deposit date: | 2023-03-08 | | Release date: | 2024-04-17 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of archaeal RNA polymerase transcription elongation and Spt4/5 recruitment.
Nucleic Acids Res., 52, 2024
|
|
