8F75
 
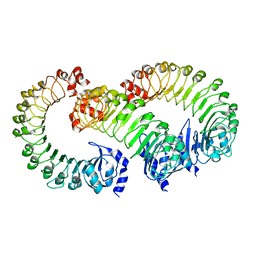 | | LRRC8A(T48D):C conformation 2 LRR focus | | Descriptor: | Volume-regulated anion channel subunit LRRC8A, Volume-regulated anion channel subunit LRRC8C | | Authors: | Kern, D.M, Brohawn, S.G. | | Deposit date: | 2022-11-18 | | Release date: | 2023-03-08 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis for assembly and lipid-mediated gating of LRRC8A:C volume-regulated anion channels.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8A41
 
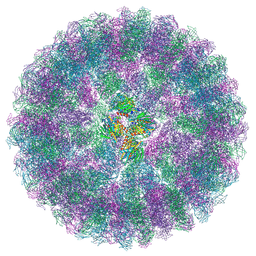 | | Nudaurelia capensis omega virus procapsid at pH7.6 (insect cell expressed VLPs) | | Descriptor: | p70 | | Authors: | Castells-Graells, R, Hesketh, E.L, Johnson, J.E, Ranson, N.A, Lawson, D.M, Lomonossoff, G.P. | | Deposit date: | 2022-06-10 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.88 Å) | | Cite: | Nudaurelia capensis omega virus maturation intermediate captured at pH5.9 (insect cell expressed VLPs)
To be published
|
|
8A3C
 
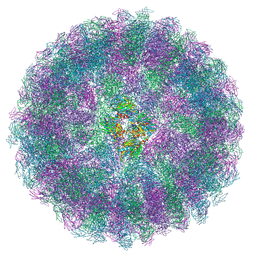 | | Nudaurelia capensis omega virus maturation intermediate captured at pH5.9 (insect cell expressed VLPs) | | Descriptor: | p70 | | Authors: | Castells-Graells, R, Hesketh, E.L, Johnson, J.E, Ranson, N.A, Lawson, D.M, Lomonossoff, G.P. | | Deposit date: | 2022-06-08 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Nudaurelia capensis omega virus maturation intermediate captured at pH5.9 (insect cell expressed VLPs)
To be published
|
|
8A6J
 
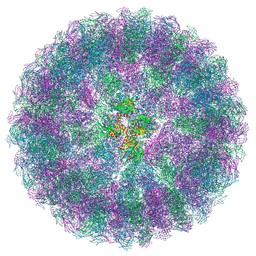 | | Nudaurelia capensis omega virus maturation intermediate captured at pH6.25 (insect cell expressed VLPs) | | Descriptor: | p70 | | Authors: | Castells-Graells, R, Hesketh, E.L, Johnson, J.E, Ranson, N.A, Lawson, D.M, Lomonossoff, G.P. | | Deposit date: | 2022-06-17 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Decoding virus maturation with cryo-EM structures of intermediates
To be published
|
|
8FX5
 
 | | Human M4 muscarinic acetylcholine receptor complex with Gi1 and xanomeline | | Descriptor: | Antibody fragment scFv16, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Vuckovic, Z, Mobbs, J.I, Glukhova, A, Sexton, P.M, Danev, R, Thal, D.M. | | Deposit date: | 2023-01-23 | | Release date: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | Xanomeline displays concomitant orthosteric and allosteric binding modes at the M 4 mAChR.
Nat Commun, 14, 2023
|
|
8FKJ
 
 | | Yeast ATP Synthase in conformation-3, at pH 6 | | Descriptor: | ATP synthase protein 8, ATP synthase subunit 4, mitochondrial, ... | | Authors: | Sharma, S, Patel, H, Luo, M, Mueller, D.M, Liao, M. | | Deposit date: | 2022-12-21 | | Release date: | 2024-01-24 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Conformational ensemble of yeast ATP synthase at low pH reveals unique intermediates and plasticity in F 1 -F o coupling.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8FL8
 
 | | Yeast ATP Synthase structure in presence of MgATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase protein 8, ... | | Authors: | Sharma, S, Patel, H, Luo, M, Mueller, D.M, Liao, M. | | Deposit date: | 2022-12-21 | | Release date: | 2024-01-17 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Conformational ensemble of yeast ATP synthase at low pH reveals unique intermediates and plasticity in F 1 -F o coupling.
Nat.Struct.Mol.Biol., 31, 2024
|
|
2CHA
 
 | |
8A3N
 
 | | Geissoschizine synthase from Catharanthus roseus - binary complex with NADP+ | | Descriptor: | Geissoschizine synthase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ZINC ION | | Authors: | Langley, C, Tatsis, E, Hong, B, Nakamura, Y, Kamileen, M.O, Paetz, C, Stevenson, C.E.M, Basquin, J, Lawson, D.M, Caputi, L, O'Connor, S.E. | | Deposit date: | 2022-06-08 | | Release date: | 2022-10-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Expansion of the Catalytic Repertoire of Alcohol Dehydrogenases in Plant Metabolism.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7Z6C
 
 | | Crystal structure of human Dihydroorotate Dehydrogenase in complex with the inhibitor 2-Hydroxy-N-(2-ispropyl-5-methyl-4-phenoxyphenyl)pyrazolo[1,5-a]pyridine-3-carboxamide. | | Descriptor: | ACETATE ION, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Alberti, M, Lolli, M.L, Boschi, D, Sainas, S, Rizzi, M, Ferraris, D.M, Miggiano, R. | | Deposit date: | 2022-03-11 | | Release date: | 2022-10-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Targeting Acute Myelogenous Leukemia Using Potent Human Dihydroorotate Dehydrogenase Inhibitors Based on the 2-Hydroxypyrazolo[1,5- a ]pyridine Scaffold: SAR of the Aryloxyaryl Moiety.
J.Med.Chem., 65, 2022
|
|
8AC6
 
 | | Nudaurelia capensis omega virus maturation intermediate captured at pH5.6 (insect cell expressed VLPs): medium class from symmetry expansion | | Descriptor: | p70 | | Authors: | Castells-Graells, R, Hesketh, E.L, Johnson, J.E, Ranson, N.A, Lawson, D.M, Lomonossoff, G.P. | | Deposit date: | 2022-07-05 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Decoding virus maturation with cryo-EM structures of intermediates
To be published
|
|
8AAY
 
 | | Nudaurelia capensis omega virus maturation intermediate captured at pH5.6 (insect cell expressed VLPs): small class from symmetry expansion | | Descriptor: | p70 | | Authors: | Castells-Graells, R, Hesketh, E.L, Johnson, J.E, Ranson, N.A, Lawson, D.M, Lomonossoff, G.P. | | Deposit date: | 2022-07-04 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Decoding virus maturation with cryo-EM structures of intermediates
To be published
|
|
8ACH
 
 | | Nudaurelia capensis omega virus maturation intermediate captured at pH5.6 (insect cell expressed VLPs): large class from symmetry expansion | | Descriptor: | p70 | | Authors: | Castells-Graells, R, Hesketh, E.L, Johnson, J.E, Ranson, N.A, Lawson, D.M, Lomonossoff, G.P. | | Deposit date: | 2022-07-05 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Decoding virus maturation with cryo-EM structures of intermediates
To be published
|
|
3L25
 
 | | Crystal structure of Zaire Ebola VP35 interferon inhibitory domain bound to 8 bp dsRNA | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Leung, D.W, Prins, K.C, Borek, D.M, Farahbakhsh, M, Tufariello, J.M, Ramanan, P, Nix, J.C, Helgeson, L.A, Otwinowski, Z, Honzatko, R.B, Basler, C.F, Amarasinghe, G.K. | | Deposit date: | 2009-12-14 | | Release date: | 2010-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for dsRNA recognition and interferon antagonism by Ebola VP35.
Nat.Struct.Mol.Biol., 17, 2010
|
|
7P9K
 
 | | BrxU, GmrSD-family Type IV restriction enzyme | | Descriptor: | CHLORIDE ION, DUF262 domain-containing protein, GLYCEROL, ... | | Authors: | Picton, D.M, Luyten, Y, Morgan, R.D, Nelson, A, Smith, D.L, Dryden, D.T.F, Hinton, J.C.D, Blower, T.R. | | Deposit date: | 2021-07-27 | | Release date: | 2021-12-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | The phage defence island of a multidrug resistant plasmid uses both BREX and type IV restriction for complementary protection from viruses.
Nucleic Acids Res., 49, 2021
|
|
7P9M
 
 | | BrxU, GmrSD-family Type IV restriction enzyme | | Descriptor: | CHLORIDE ION, DUF262 domain-containing protein, SULFATE ION | | Authors: | Picton, D.M, Luyten, Y, Morgan, R.D, Nelson, A, Smith, D.L, Dryden, D.T.F, Hinton, J.C.D, Blower, T.R. | | Deposit date: | 2021-07-27 | | Release date: | 2021-12-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The phage defence island of a multidrug resistant plasmid uses both BREX and type IV restriction for complementary protection from viruses.
Nucleic Acids Res., 49, 2021
|
|
5QHM
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of human FAM83B in complex with OX-145 | | Descriptor: | 1,2-ETHANEDIOL, 1-methyl-3-oxidanyl-pyridine-2-thione, Protein FAM83B | | Authors: | Pinkas, D.M, Bufton, J.C, Fox, A.E, Talon, R, Krojer, T, Douangamath, A, Collins, P, Zhang, R, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Bullock, A.N. | | Deposit date: | 2018-05-18 | | Release date: | 2018-12-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5OY6
 
 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with cyclical inhibitor OD36. | | Descriptor: | 1,2-ETHANEDIOL, Activin receptor type-1, cyclical inhibitor OD36 | | Authors: | Williams, E.P, Pinkas, D.M, Krojer, T, Kupinska, K, Mahajan, P, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2017-09-07 | | Release date: | 2017-09-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Establishment and characterization of endothelial colony forming cells as a surrogate model for Fibrodysplasia Ossificans Progressiva
To Be Published
|
|
5QHN
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of human FAM83B in complex with FMOPL000622a | | Descriptor: | 1,2-ETHANEDIOL, Protein FAM83B, methyl (2~{S},4~{R})-1-(furan-2-ylcarbonyl)-4-oxidanyl-pyrrolidine-2-carboxylate | | Authors: | Pinkas, D.M, Bufton, J.C, Fox, A.E, Talon, R, Krojer, T, Douangamath, A, Collins, P, Zhang, R, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Bullock, A.N. | | Deposit date: | 2018-05-18 | | Release date: | 2018-12-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QHK
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of human FAM83B in complex with FMOPL000010a | | Descriptor: | 1,2-ETHANEDIOL, IODIDE ION, Protein FAM83B, ... | | Authors: | Pinkas, D.M, Bufton, J.C, Fox, A.E, Talon, R, Krojer, T, Douangamath, A, Collins, P, Zhang, R, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Bullock, A.N. | | Deposit date: | 2018-05-18 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
3L27
 
 | | Crystal structure of Zaire Ebola VP35 interferon inhibitory domain R312A mutant | | Descriptor: | CHLORIDE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Leung, D.W, Prins, K.C, Borek, D.M, Farahbakhsh, M, Tufariello, J.M, Ramanan, P, Nix, J.C, Helgeson, L.A, Otwinowski, Z, Honzatko, R.B, Basler, C.F, Amarasinghe, G.K. | | Deposit date: | 2009-12-14 | | Release date: | 2010-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for dsRNA recognition and interferon antagonism by Ebola VP35.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3L26
 
 | | Crystal structure of Zaire Ebola VP35 interferon inhibitory domain bound to 8 bp dsRNA | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Polymerase cofactor VP35, ... | | Authors: | Leung, D.W, Prins, K.C, Borek, D.M, Farahbakhsh, M, Tufariello, J.M, Ramanan, P, Nix, J.C, Helgeson, L.A, Otwinowski, Z, Honzatko, R.B, Basler, C.F, Amarasinghe, G.K. | | Deposit date: | 2009-12-14 | | Release date: | 2010-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for dsRNA recognition and interferon antagonism by Ebola VP35.
Nat.Struct.Mol.Biol., 17, 2010
|
|
5QHQ
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of human FAM83B in complex with FMOPL000574a | | Descriptor: | 1,2-ETHANEDIOL, 1-[(4-fluorophenyl)methyl]benzimidazole, Protein FAM83B | | Authors: | Pinkas, D.M, Bufton, J.C, Fox, A.E, Talon, R, Krojer, T, Douangamath, A, Collins, P, Zhang, R, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Bullock, A.N. | | Deposit date: | 2018-05-18 | | Release date: | 2018-12-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QHI
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of human FAM83B in complex with FMOPL000271a | | Descriptor: | 1,2-ETHANEDIOL, IODIDE ION, Protein FAM83B, ... | | Authors: | Pinkas, D.M, Bufton, J.C, Fox, A.E, Talon, R, Krojer, T, Douangamath, A, Collins, P, Zhang, R, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Bullock, A.N. | | Deposit date: | 2018-05-18 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
7P2W
 
 | | E.coli GyrB24 with inhibitor LMD92 (EBL2682) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[[3,4-bis(chloranyl)-5-methyl-1H-pyrrol-2-yl]carbonylamino]-4-[(3-carboxyphenyl)methoxy]-1,3-benzothiazole-6-carboxylic acid, DNA gyrase subunit B, ... | | Authors: | Stevenson, C.E.M, Lawson, D.M, Maxwell, A.M, Henderson, S.R, Kikelj, D, Durcik, M, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P. | | Deposit date: | 2021-07-06 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery and Hit-to-Lead Optimization of Benzothiazole Scaffold-Based DNA Gyrase Inhibitors with Potent Activity against Acinetobacter baumannii and Pseudomonas aeruginosa.
J.Med.Chem., 66, 2023
|
|
