1XLA
 
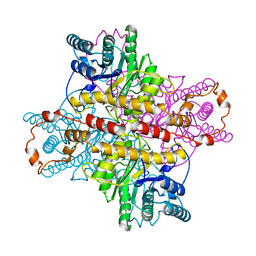 | |
1XLI
 
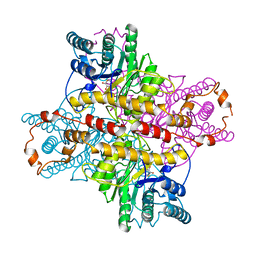 | | MECHANISM FOR ALDOSE-KETOSE INTERCONVERSION BY D-XYLOSE ISOMERASE INVOLVING RING OPENING FOLLOWED BY A 1,2-HYDRIDE SHIFT | | Descriptor: | 5-thio-alpha-D-glucopyranose, D-XYLOSE ISOMERASE, MANGANESE (II) ION | | Authors: | Collyer, C.A, Henrick, K, Blow, D.M. | | Deposit date: | 1991-10-09 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism for aldose-ketose interconversion by D-xylose isomerase involving ring opening followed by a 1,2-hydride shift.
J.Mol.Biol., 212, 1990
|
|
1XLF
 
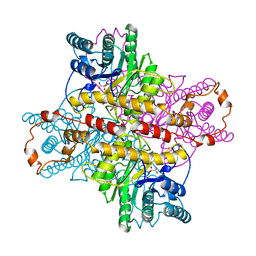 | | MECHANISM FOR ALDOSE-KETOSE INTERCONVERSION BY D-XYLOSE ISOMERASE INVOLVING RING OPENING FOLLOWED BY A 1,2-HYDRIDE SHIFT | | Descriptor: | D-XYLOSE ISOMERASE, D-gluconic acid, MANGANESE (II) ION | | Authors: | Collyer, C.A, Henrick, K, Blow, D.M. | | Deposit date: | 1991-10-09 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism for aldose-ketose interconversion by D-xylose isomerase involving ring opening followed by a 1,2-hydride shift.
J.Mol.Biol., 212, 1990
|
|
1XLC
 
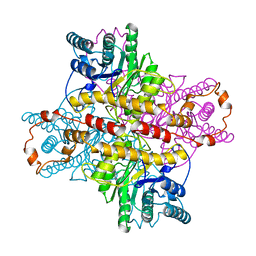 | | MECHANISM FOR ALDOSE-KETOSE INTERCONVERSION BY D-XYLOSE ISOMERASE INVOLVING RING OPENING FOLLOWED BY A 1,2-HYDRIDE SHIFT | | Descriptor: | D-XYLOSE ISOMERASE, MAGNESIUM ION, Xylitol | | Authors: | Collyer, C.A, Henrick, K, Blow, D.M. | | Deposit date: | 1991-10-09 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism for aldose-ketose interconversion by D-xylose isomerase involving ring opening followed by a 1,2-hydride shift.
J.Mol.Biol., 212, 1990
|
|
1XLL
 
 | | MECHANISM FOR ALDOSE-KETOSE INTERCONVERSION BY D-XYLOSE ISOMERASE INVOLVING RING OPENING FOLLOWED BY A 1,2-HYDRIDE SHIFT | | Descriptor: | D-XYLOSE ISOMERASE, ZINC ION | | Authors: | Collyer, C.A, Henrick, K, Blow, D.M. | | Deposit date: | 1991-10-09 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism for aldose-ketose interconversion by D-xylose isomerase involving ring opening followed by a 1,2-hydride shift.
J.Mol.Biol., 212, 1990
|
|
4U6H
 
 | | Vaccinia L1/M12B9-Fab complex | | Descriptor: | Heavy chain of murine anti-vaccinia L1 IgG2a antibody M12B9, Light chain of murine anti-vaccinia L1 IgG2a antibody M12B9, Protein L1 | | Authors: | Matho, M.H, Schlossman, A, Zajonc, D.M. | | Deposit date: | 2014-07-29 | | Release date: | 2014-08-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Potent neutralization of vaccinia virus by divergent murine antibodies targeting a common site of vulnerability in l1 protein.
J.Virol., 88, 2014
|
|
4TXE
 
 | | ScCTS1 in complex with compound 5 | | Descriptor: | (2S)-1-(2,3-dihydro-1H-inden-2-ylamino)-3-(3,4-dimethylphenoxy)propan-2-ol, Endochitinase | | Authors: | Schuettelkopf, A.W, van Aalten, D.M.F. | | Deposit date: | 2014-07-03 | | Release date: | 2014-08-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Screening-based discovery of Aspergillus fumigatus plant-type chitinase inhibitors
FEBS Lett, 588, 2014
|
|
4TX6
 
 | | AfChiA1 in complex with compound 1 | | Descriptor: | 3-(2-methoxyphenyl)-6-methyl[1,2]oxazolo[5,4-d]pyrimidin-4(5H)-one, Class III chitinase ChiA1, PHOSPHATE ION | | Authors: | van Aalten, D.M.F. | | Deposit date: | 2014-07-02 | | Release date: | 2014-08-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Screening-based discovery of Aspergillus fumigatus plant-type chitinase inhibitors
FEBS Lett, 588, 2014
|
|
4UIJ
 
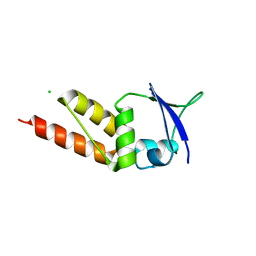 | | Crystal structure of the BTB domain of KCTD13 | | Descriptor: | BTB/POZ DOMAIN-CONTAINING ADAPTER FOR CUL3-MEDIATED RHOA DEGRADATION PROTEIN 1, CHLORIDE ION | | Authors: | Pinkas, D.M, Sanvitale, C.E, Sorell, F.J, Solcan, N, Goubin, S, Canning, P, Williams, E, Chaikuad, A, Dixon Clarke, S.E, Tallant, C, Fonseca, M, Chalk, R, Doutch, J, Krojer, T, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2015-03-30 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural complexity in the KCTD family of Cullin3-dependent E3 ubiquitin ligases.
Biochem. J., 474, 2017
|
|
1MWN
 
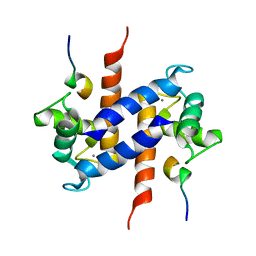 | | Solution NMR structure of S100B bound to the high-affinity target peptide TRTK-12 | | Descriptor: | CALCIUM ION, F-actin capping protein alpha-1 subunit, S-100 protein, ... | | Authors: | Inman, K.G, Yang, R, Rustandi, R.R, Miller, K.E, Baldisseri, D.M, Weber, D.J. | | Deposit date: | 2002-09-30 | | Release date: | 2002-12-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of S100B bound to the high-affinity target peptide TRTK-12
J.Mol.Biol., 324, 2002
|
|
4UNO
 
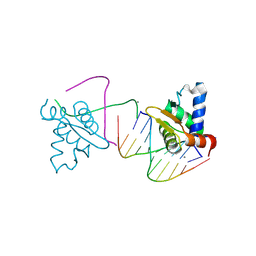 | | Crystal structure of the ETS domain of human ETV5 in complex with DNA | | Descriptor: | 5'-D(*AP*CP*CP*GP*GP*AP*AP*GP*TP*GP)-3', 5'-D(*AP*CP*TP*TP*CP*CP*GP*GP*TP*CP)-3', CALCIUM ION, ... | | Authors: | Newman, J.A, Aitkenhead, H, Cooper, C.D.O, Pinkas, D.M, Shrestha, L, Burgess-Brown, N, Kopec, J, Fitzpatrick, F, Tallant, C, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Gileadi, O. | | Deposit date: | 2014-05-29 | | Release date: | 2014-06-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of the Ets Domains of Transcription Factors Etv1, Etv4, Etv5 and Fev: Determinants of DNA Binding and Redox Regulation by Disulfide Bond Formation.
J.Biol.Chem., 290, 2015
|
|
4V11
 
 | | Structure of Synaptotagmin-1 with SV2A peptide phosphorylated at Thr84 | | Descriptor: | CALCIUM ION, GLYCEROL, SYNAPTIC VESICLE GLYCOPROTEIN 2A, ... | | Authors: | Zhang, N, Gordon, S.L, Fritsch, M.J, Esoof, N, Campbell, D, Gourlay, R, Velupillai, S, Macartney, T, Peggie, M, vanAalten, D.M.F, Cousin, M.A, Alessi, D.R. | | Deposit date: | 2014-09-22 | | Release date: | 2015-02-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Phosphorylation of Synaptic Vesicle Protein 2A at Thr84 by Casein Kinase 1 Family Kinases Controls the Specific Retrieval of Synaptotagmin-1.
J.Neurosci., 35, 2015
|
|
4QAG
 
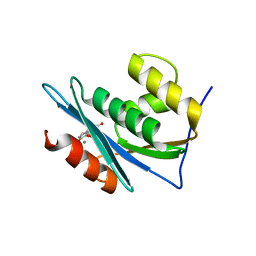 | | Structure of a dihydroxycoumarin active-site inhibitor in complex with the RNASE H domain of HIV-1 reverse transcriptase | | Descriptor: | (7,8-dihydroxy-2-oxo-2H-chromen-4-yl)acetic acid, MANGANESE (II) ION, Reverse transcriptase/ribonuclease H | | Authors: | Himmel, D.M, Ho, W.C, Arnold, E. | | Deposit date: | 2014-05-04 | | Release date: | 2014-06-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.712 Å) | | Cite: | Structure of a Dihydroxycoumarin Active-Site Inhibitor in Complex with the RNase H Domain of HIV-1 Reverse Transcriptase and Structure-Activity Analysis of Inhibitor Analogs.
J.Mol.Biol., 426, 2014
|
|
1EST
 
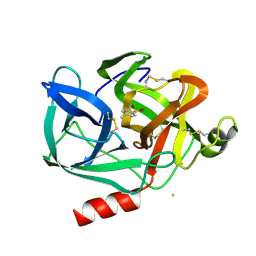 | |
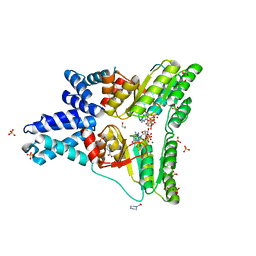
1OKK
 
 | |
1V45
 
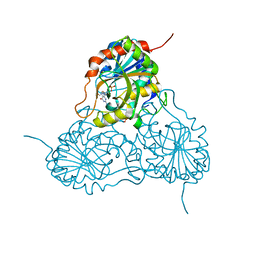 | | Crystal Structure of human PNP complexed with 3-deoxyguanosine | | Descriptor: | 9-(3-DEOXY-BETA-D-RIBOFURANOSYL)GUANINE, Purine nucleoside phosphorylase, SULFATE ION | | Authors: | Canduri, F, Dos Santos, D.M, Silva, R.G, Mendes, M.A, Palma, M.S, De Azevedo Jr, W.F, Basso, L.A, Santos, D.S. | | Deposit date: | 2003-11-10 | | Release date: | 2004-12-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structure of human PNP complexed with ligands.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
3U7C
 
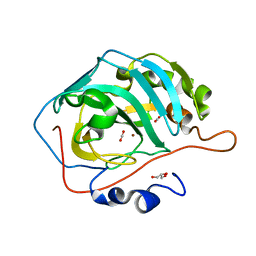 | | crystal structure of the V143I mutant of human carbonic anhydrase II | | Descriptor: | BICARBONATE ION, CARBON DIOXIDE, Carbonic anhydrase 2, ... | | Authors: | West, D.M, Kim, C.U, Robbins, A.H, Mckenna, R. | | Deposit date: | 2011-10-13 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | crystal structure of the V143I mutant of human carbonic anhydrase II
To be Published
|
|
6EOU
 
 | |
5XKS
 
 | |
6TB7
 
 | |
5X61
 
 | | Crystal structure of Acetylcholinesterase Catalytic Subunit of the Malaria Vector Anopheles Gambiae, 3.4 A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AMMONIUM ION, ... | | Authors: | Han, Q, Robinson, H, Ding, H, Wong, D.M, Lam, P.C.H, Totrov, M.M, Carlier, P.R, Li, J. | | Deposit date: | 2017-02-20 | | Release date: | 2017-03-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structure of acetylcholinesterase catalytic subunits of the malaria vector Anopheles gambiae
Insect Sci., 2017
|
|
5X6U
 
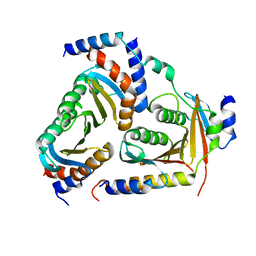 | | Crystal structure of human heteropentameric complex | | Descriptor: | Ragulator complex protein LAMTOR1, Ragulator complex protein LAMTOR2, Ragulator complex protein LAMTOR3, ... | | Authors: | Yonehara, R, Nada, S, Nakai, T, Nakai, M, Kitamura, A, Ogawa, A, Nakatsumi, H, Nakayama, K.I, Li, S, Standley, D.M, Yamashita, E, Nakagawa, A, Okada, M. | | Deposit date: | 2017-02-23 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the assembly of the Ragulator-Rag GTPase complex.
Nat Commun, 8, 2017
|
|
6TF2
 
 | |
5JLT
 
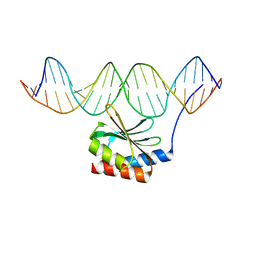 | | The crystal structure of the bacteriophage T4 MotA C-terminal domain in complex with dsDNA reveals a novel protein-DNA recognition motif | | Descriptor: | DNA (5'-D(*GP*AP*AP*GP*CP*TP*TP*TP*GP*CP*TP*TP*AP*AP*TP*AP*AP*TP*CP*CP*AP*C)-3'), DNA (5'-D(*GP*TP*GP*GP*AP*TP*TP*AP*TP*TP*AP*AP*GP*CP*AP*AP*AP*GP*CP*TP*TP*C)-3'), Middle transcription regulatory protein motA | | Authors: | Cuypers, M.G, Robertson, R.M, Knipling, L, Hinton, D.M, White, S.W. | | Deposit date: | 2016-04-27 | | Release date: | 2017-05-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.955 Å) | | Cite: | The phage T4 MotA transcription factor contains a novel DNA binding motif that specifically recognizes modified DNA.
Nucleic Acids Res., 46, 2018
|
|
6T3E
 
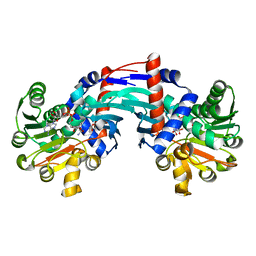 | | Structure of Thermococcus litoralis Delta(1)-pyrroline-2-carboxylate reductase in complex with NADH and L-proline | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, DELTA1-pyrroline-2-carboxylate reductase, PROLINE | | Authors: | Ferraris, D.M, Miggiano, R, Ferrario, E, Rizzi, M. | | Deposit date: | 2019-10-10 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Thermococcus litoralis Delta1-pyrroline-2-carboxylate reductase in complex with NADH and L-proline.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
