2LZZ
 
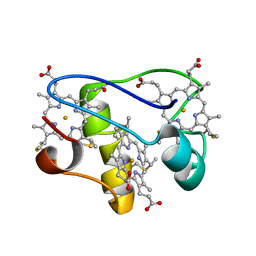 | | Solution structure of a mutant of the triheme cytochrome PpcA from Geobacter sulfurreducens sheds light on the role of the conserved aromatic residue F15 | | Descriptor: | Cytochrome c, 3 heme-binding sites, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Dantas, J.M, Morgado, L, Turner, D.L, Salgueiro, C.A. | | Deposit date: | 2012-10-12 | | Release date: | 2013-01-30 | | Last modified: | 2013-03-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a mutant of the triheme cytochrome PpcA from Geobacter sulfurreducens sheds light on the role of the conserved aromatic residue F15.
Biochim.Biophys.Acta, 1827, 2013
|
|
2M22
 
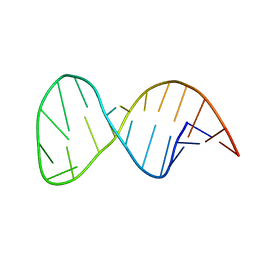 | | Solution structure of the helix II template boundary element from Tetrahymena telomerase RNA | | Descriptor: | 5'-R(*GP*GP*CP*AP*GP*AP*UP*CP*UP*GP*UP*AP*AP*UP*AP*GP*AP*AP*CP*UP*GP*CP*C)-3' | | Authors: | Cash, D.D, Richards, R.J, Theimer, C.A, Finger, D.L, Feigon, J. | | Deposit date: | 2012-12-11 | | Release date: | 2013-03-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the helix II template boundary element from Tetrahymena telomerase RNA
Nucleic Acids Res., 34, 2006
|
|
2MGX
 
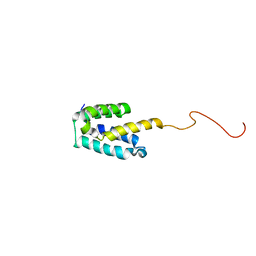 | | NMR structure of SRA1p C-terminal domain | | Descriptor: | Steroid receptor RNA activator 1 | | Authors: | Bilinovich, S.M, Davis, C.M, Morris, D.L, Ray, L.A, Prokop, J.W, Buchan, G.J, Leeper, T.C. | | Deposit date: | 2013-11-10 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The C-Terminal Domain of SRA1p Has a Fold More Similar to PRP18 than to an RRM and Does Not Directly Bind to the SRA1 RNA STR7 Region.
J.Mol.Biol., 426, 2014
|
|
2MZ9
 
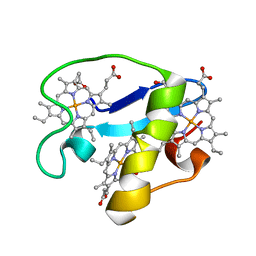 | | Solution structure of oxidized triheme cytochrome PpcA from Geobacter sulfurreducens | | Descriptor: | HEME C, PpcA | | Authors: | Morgado, L, Bruix, M, Pokkuluri, R, Salgueiro, C.A, Turner, D.L. | | Deposit date: | 2015-02-08 | | Release date: | 2016-02-10 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Redox- and pH-linked conformational changes in triheme cytochrome PpcA from Geobacter sulfurreducens.
Biochem. J., 474, 2017
|
|
1Z64
 
 | | NMR Solution Structure of Pleurocidin in DPC Micelles | | Descriptor: | Pleruocidin | | Authors: | Syvitski, R.T, Burton, I, Mattatall, N.R, Douglas, S.E, Jakeman, D.L. | | Deposit date: | 2005-03-21 | | Release date: | 2005-04-12 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of the antimicrobial peptide pleurocidin from winter flounder.
Biochemistry, 44, 2005
|
|
2MS3
 
 | |
1ZI9
 
 | | Crystal Structure Analysis of the dienelactone hydrolase (E36D, C123S) mutant- 1.5 A | | Descriptor: | Carboxymethylenebutenolidase, GLYCEROL, SULFATE ION | | Authors: | Kim, H.-K, Liu, J.-W, Carr, P.D, Ollis, D.L. | | Deposit date: | 2005-04-27 | | Release date: | 2005-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Following directed evolution with crystallography: structural changes observed in changing the substrate specificity of dienelactone hydrolase.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1ZJ4
 
 | | Crystal Structure Analysis of the dienelactone hydrolase mutant (E36D, C123S) bound with the PMS moiety of the protease inhibitor, Phenylmethylsulfonyl fluoride (PMSF)- 1.7 A | | Descriptor: | Carboxymethylenebutenolidase, GLYCEROL, SULFATE ION | | Authors: | Kim, H.-K, Liu, J.-W, Carr, P.D, Ollis, D.L. | | Deposit date: | 2005-04-28 | | Release date: | 2005-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Following directed evolution with crystallography: structural changes observed in changing the substrate specificity of dienelactone hydrolase.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1YKJ
 
 | | A45G p-hydroxybenzoate hydroxylase with p-hydroxybenzoate bound | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOIC ACID, P-hydroxybenzoate hydroxylase, ... | | Authors: | Cole, L.J, Gatti, D.L, Entsch, B, Ballou, D.P. | | Deposit date: | 2005-01-18 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Removal of a methyl group causes global changes in p-hydroxybenzoate hydroxylase.
Biochemistry, 44, 2005
|
|
1YD2
 
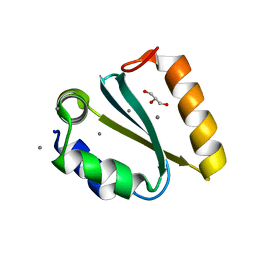 | | Crystal structure of the GIY-YIG N-terminal endonuclease domain of UvrC from Thermotoga maritima: Point mutant Y19F bound to the catalytic divalent cation | | Descriptor: | GLYCEROL, MANGANESE (II) ION, UvrABC system protein C | | Authors: | Truglio, J.J, Rhau, B, Croteau, D.L, Wang, L, Skorvaga, M, Karakas, E, DellaVecchia, M.J, Wang, H, Van Houten, B, Kisker, C. | | Deposit date: | 2004-12-23 | | Release date: | 2005-03-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into the first incision reaction during nucleotide excision repair
Embo J., 24, 2005
|
|
2NUF
 
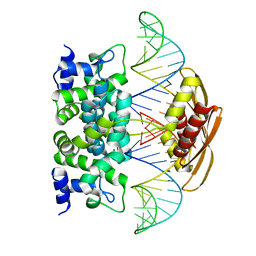 | | Crystal structure of RNase III from Aquifex aeolicus complexed with ds-RNA at 2.5-Angstrom Resolution | | Descriptor: | 28-MER, MAGNESIUM ION, Ribonuclease III | | Authors: | Gan, J.H, Shaw, G, Tropea, J.E, Waugh, D.S, Court, D.L, Ji, X. | | Deposit date: | 2006-11-09 | | Release date: | 2007-11-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A stepwise model for double-stranded RNA processing by ribonuclease III.
Mol.Microbiol., 67, 2007
|
|
2NWR
 
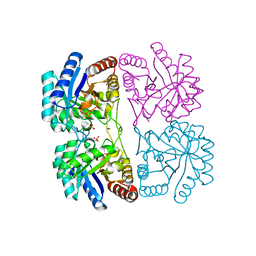 | | Crystal structure of C11N mutant of KDO8P Synthase in complex with PEP | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase, PHOSPHATE ION, PHOSPHOENOLPYRUVATE | | Authors: | Kona, F, Xu, X, Martin, P, Kuzmic, P, Gatti, D.L. | | Deposit date: | 2006-11-16 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Mechanistic Changes along an Engineered Path from Metallo to Nonmetallo 3-Deoxy-d-manno-octulosonate 8-Phosphate Synthases.
Biochemistry, 46, 2007
|
|
2NXI
 
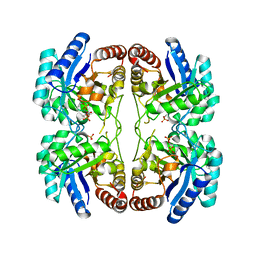 | | Structural and mechanistic changes along an engineered path from metallo to non-metallo KDO8P synthase. | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase, PHOSPHATE ION, PHOSPHOENOLPYRUVATE | | Authors: | Kona, F, Xu, X, Martin, P, Kuzmic, P, Gatti, D.L. | | Deposit date: | 2006-11-17 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and mechanistic changes along an engineered path from metallo to nonmetallo 3-deoxy-D-manno-octulosonate 8-phosphate synthases
Biochemistry, 46, 2007
|
|
1ZIY
 
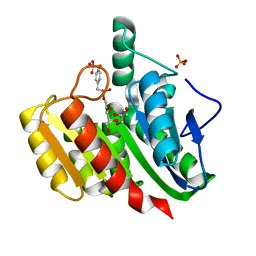 | | Crystal Structure Analysis of the dienelactone hydrolase mutant (C123S) bound with the PMS moiety of the protease inhibitor, Phenylmethylsulfonyl fluoride (PMSF)- 1.9 A | | Descriptor: | Carboxymethylenebutenolidase, GLYCEROL, SULFATE ION | | Authors: | Kim, H.-K, Liu, J.-W, Carr, P.D, Ollis, D.L. | | Deposit date: | 2005-04-27 | | Release date: | 2005-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Following directed evolution with crystallography: structural changes observed in changing the substrate specificity of dienelactone hydrolase.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2NWS
 
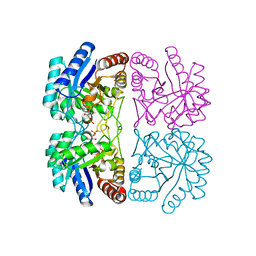 | | Structural and mechanistic changes along an engineered path from metallo to non-metallo KDO8P synthase | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase, ARABINOSE-5-PHOSPHATE, PHOSPHOENOLPYRUVATE | | Authors: | Kona, F, Xu, X, Martin, P, Kuzmic, P, Gatti, D.L. | | Deposit date: | 2006-11-16 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Mechanistic Changes along an Engineered Path from Metallo to Nonmetallo 3-Deoxy-d-manno-octulosonate 8-Phosphate Synthases.
Biochemistry, 46, 2007
|
|
1XNY
 
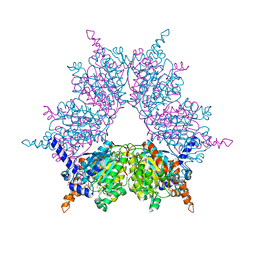 | | Biotin and propionyl-CoA bound to Acyl-CoA Carboxylase Beta Subunit from S. coelicolor (PccB) | | Descriptor: | BIOTIN, propionyl Coenzyme A, propionyl-CoA carboxylase complex B subunit | | Authors: | Diacovich, L, Mitchell, D.L, Pham, H, Gago, G, Melgar, M.M, Khosla, C, Gramajo, H, Tsai, S.-C. | | Deposit date: | 2004-10-05 | | Release date: | 2004-11-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the beta-Subunit of Acyl-CoA Carboxylase: Structure-Based Engineering of Substrate Specificity
Biochemistry, 43, 2004
|
|
2NUG
 
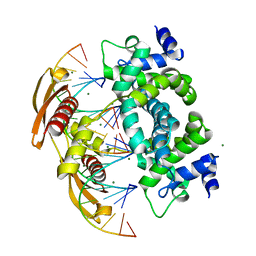 | | Crystal structure of RNase III from Aquifex aeolicus complexed with ds-RNA at 1.7-Angstrom Resolution | | Descriptor: | 5'-R(P*AP*AP*GP*GP*UP*CP*AP*UP*UP*CP*G)-3', 5'-R(P*AP*GP*UP*GP*GP*CP*CP*UP*UP*GP*C)-3', MAGNESIUM ION, ... | | Authors: | Gan, J.H, Shaw, G, Tropea, J.E, Waugh, D.S, Court, D.L, Ji, X. | | Deposit date: | 2006-11-09 | | Release date: | 2007-11-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A stepwise model for double-stranded RNA processing by ribonuclease III.
Mol.Microbiol., 67, 2007
|
|
1YXN
 
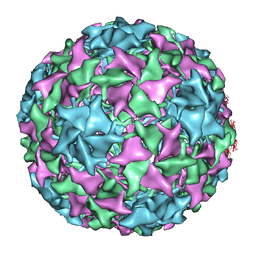 | | Pseudo-atomic model of a fiberless isometric variant of bacteriophage phi29 | | Descriptor: | Major head protein | | Authors: | Morais, M.C, Choi, K.H, Koti, J.S, Chipman, P.R, Anderson, D.L, Rossmann, M.G. | | Deposit date: | 2005-02-22 | | Release date: | 2005-04-26 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Conservation of the Capsid Structure in Tailed dsDNA Bacteriophages: the Pseudoatomic Structure of phi29
Mol.Cell, 18, 2005
|
|
1YYK
 
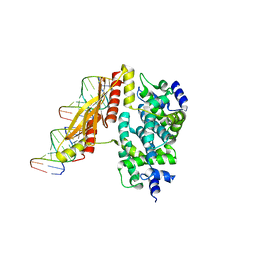 | | Crystal structure of RNase III from Aquifex Aeolicus complexed with double-stranded RNA at 2.5-angstrom resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-R(*CP*GP*CP*GP*AP*AP*UP*UP*CP*GP*CP*G)-3', Ribonuclease III | | Authors: | Gan, J, Tropea, J.E, Austin, B.P, Court, D.L, Waugh, D.S, Ji, X. | | Deposit date: | 2005-02-25 | | Release date: | 2005-11-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Intermediate states of ribonuclease III in complex with double-stranded RNA
Structure, 13, 2005
|
|
2O94
 
 | |
1YYO
 
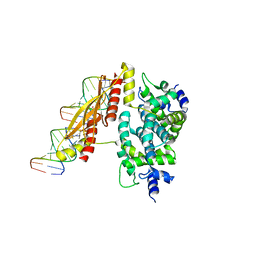 | | Crystal structure of RNase III mutant E110K from Aquifex aeolicus complexed with double-stranded RNA at 2.9-Angstrom Resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-R(*CP*GP*CP*GP*AP*AP*UP*UP*CP*GP*CP*G)-3', Ribonuclease III | | Authors: | Gan, J, Tropea, J.E, Austin, B.P, Court, D.L, Waugh, D.S, Ji, X. | | Deposit date: | 2005-02-25 | | Release date: | 2005-11-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Intermediate states of ribonuclease III in complex with double-stranded RNA
Structure, 13, 2005
|
|
1YZ9
 
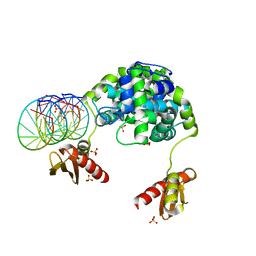 | | Crystal structure of RNase III mutant E110Q from Aquifex aeolicus complexed with double stranded RNA at 2.1-Angstrom Resolution | | Descriptor: | 5'-R(*CP*GP*AP*AP*CP*UP*UP*CP*GP*CP*G)-3', Ribonuclease III, SULFATE ION | | Authors: | Gan, J, Tropea, J.E, Austin, B.P, Court, D.L, Waugh, D.S, Ji, X. | | Deposit date: | 2005-02-28 | | Release date: | 2005-11-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Intermediate states of ribonuclease III in complex with double-stranded RNA
Structure, 13, 2005
|
|
2P4X
 
 | |
2OLD
 
 | | Bence Jones KWR Protein- Immunoglobulin Light Chain Dimer, P3(2)21 Crystal Form | | Descriptor: | Bence Jones KWR Protein - Immunoglobulin Light Chain, PHENOL, PHOSPHATE ION | | Authors: | Makino, D.L, Larson, S.B, McPherson, A. | | Deposit date: | 2007-01-19 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Bence Jones KWR protein structures determined by X-ray crystallography.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2OMN
 
 | | Bence Jones KWR Protein- Immunoglobulin Light Chain Dimer, P4(3)2(1)2 Crystal Form | | Descriptor: | Bence Jones KWR Protein - Immunoglobulin Light Chain, PHENOL, SULFATE ION | | Authors: | Makino, D.L, Larson, S.B, McPherson, A. | | Deposit date: | 2007-01-22 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bence Jones KWR protein structures determined by X-ray crystallography.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
