1GGV
 
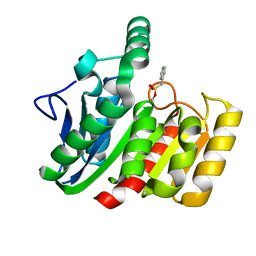 | | CRYSTAL STRUCTURE OF THE C123S MUTANT OF DIENELACTONE HYDROLASE (DLH) BOUND WITH THE PMS MOIETY OF THE PROTEASE INHIBITOR, PHENYLMETHYLSULFONYL FLUORIDE (PMSF) | | Descriptor: | DIENELACTONE HYDROLASE | | Authors: | Robinson, A, Edwards, K.J, Carr, P.D, Barton, J.D, Ewart, G.D, Ollis, D.L. | | Deposit date: | 2000-09-27 | | Release date: | 2000-12-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the C123S mutant of dienelactone hydrolase (DLH) bound with the PMS moiety of the protease inhibitor phenylmethylsulfonyl fluoride (PMSF).
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1GMY
 
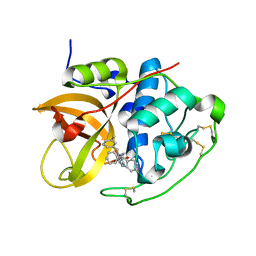 | | Cathepsin B complexed with dipeptidyl nitrile inhibitor | | Descriptor: | 2-AMINOETHANIMIDIC ACID, 3-METHYLPHENYLALANINE, CATHEPSIN B, ... | | Authors: | Greenspan, P.D, Clark, K.L, Tommasi, R.A, Cowen, S.D, McQuire, L.W, Farley, D.L, van Duzer, J.H, Goldberg, R.L, Zhou, H, Du, Z, Fitt, J.J, Coppa, D.E, Fang, Z, Macchia, W, Zhu, L, Capparelli, M.P, Goldstein, R, Wigg, A.M, Doughty, J.R, Bohacek, R.S, Knap, A.K. | | Deposit date: | 2001-09-25 | | Release date: | 2002-09-19 | | Last modified: | 2017-07-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of Dipeptidyl Nitriles as Potent and Selective Inhibitors of Cathepsin B Through Structure-Based Drug Design
J.Med.Chem., 44, 2001
|
|
1GNK
 
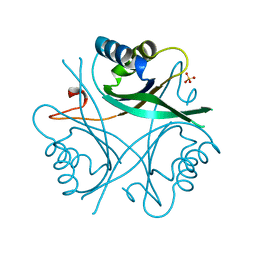 | | GLNK, A SIGNAL PROTEIN FROM E. COLI | | Descriptor: | PROTEIN (GLNK), SULFATE ION | | Authors: | Xu, Y, Cheah, E, Carr, P.D, Vanheeswijk, W.C, Westerhoff, H.V, Vasudevan, S.G, Ollis, D.L. | | Deposit date: | 1998-07-14 | | Release date: | 1999-07-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | GlnK, a PII-homologue: structure reveals ATP binding site and indicates how the T-loops may be involved in molecular recognition.
J.Mol.Biol., 282, 1998
|
|
1DIN
 
 | |
1IMW
 
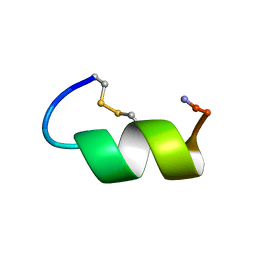 | | Peptide Antagonist of IGFBP-1 | | Descriptor: | IGFBP-1 antagonist | | Authors: | Lowman, H.B, Chen, Y.M, Skelton, N.J, Mortensen, D.L, Tomlinson, E.E, Sadick, M.D, Robinson, I.C, Clark, R.G. | | Deposit date: | 2001-05-11 | | Release date: | 2001-05-30 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of a phage display-derived peptide that binds to insulin-like growth factor binding protein 1.
Biochemistry, 40, 2001
|
|
1LCA
 
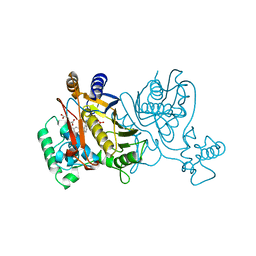 | | LACTOBACILLUS CASEI THYMIDYLATE SYNTHASE TERNARY COMPLEX WITH DUMP AND CB3717 | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, THYMIDYLATE SYNTHASE | | Authors: | Birdsall, D.L, Finer-Moore, J, Stroud, R.M. | | Deposit date: | 1995-06-22 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Refined structures of substrate-bound and phosphate-bound thymidylate synthase from Lactobacillus casei.
J.Mol.Biol., 232, 1993
|
|
3HX4
 
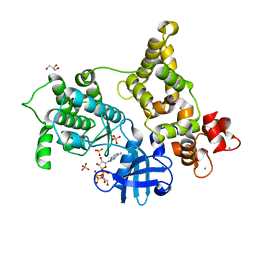 | | Crystal structure of CDPK1 of Toxoplasma gondii, TGME49_101440, in presence of calcium | | Descriptor: | CALCIUM ION, Calmodulin-domain protein kinase 1, GLYCEROL, ... | | Authors: | Wernimont, A.K, Artz, J.D, Finnerty, P, Xiao, T, He, H, MacKenzie, F, Sinestera, G, Hassani, A.A, Wasney, G, Vedadi, M, Lourido, S, Bochkarev, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Sibley, D.L, Hui, R, Lin, Y.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-06-19 | | Release date: | 2009-07-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of apicomplexan calcium-dependent protein kinases reveal mechanism of activation by calcium.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3IAV
 
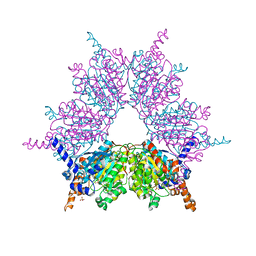 | | Propionyl-CoA Carboxylase Beta Subunit, D422V | | Descriptor: | Propionyl-CoA carboxylase complex B subunit, SULFATE ION | | Authors: | Diacovich, L, Arabolaza, A, Shillito, E.M, Lin, T.-W, Mitchell, D.L, Pham, H, Melgar, M.M. | | Deposit date: | 2009-07-14 | | Release date: | 2010-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures and mutational analyses of acyl-CoA carboxylase beta subunit of Streptomyces coelicolor.
Biochemistry, 49, 2010
|
|
3IBB
 
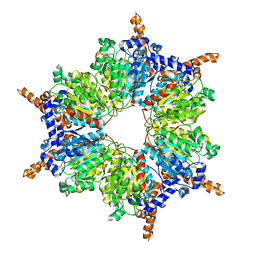 | | Propionyl-CoA Carboxylase Beta Subunit, D422A | | Descriptor: | Propionyl-CoA carboxylase complex B subunit | | Authors: | Diacovich, L, Arabolaza, A, Shillito, E.M, Lin, T.-W, Mitchell, D.L, Pham, H, Melgar, M.M. | | Deposit date: | 2009-07-15 | | Release date: | 2010-06-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structures and mutational analyses of acyl-CoA carboxylase beta subunit of Streptomyces coelicolor.
Biochemistry, 49, 2010
|
|
3IB9
 
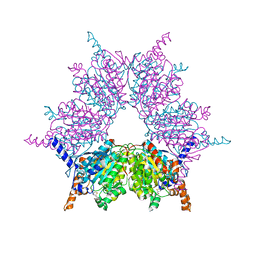 | | Propionyl-CoA Carboxylase Beta Subunit, D422L | | Descriptor: | BIOTIN, Propionyl-CoA carboxylase complex B subunit, SULFATE ION | | Authors: | Diacovich, L, Arabolaza, A, Shillito, E.M, Lin, T.-W, Mitchell, D.L, Pham, H, Melgar, M.M. | | Deposit date: | 2009-07-15 | | Release date: | 2010-06-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures and mutational analyses of acyl-CoA carboxylase beta subunit of Streptomyces coelicolor.
Biochemistry, 49, 2010
|
|
1GJE
 
 | | Peptide Antagonist of IGFBP-1, Minimized Average Structure | | Descriptor: | IGFBP-1 antagonist | | Authors: | Lowman, H.B, Chen, Y.M, Skelton, N.J, Mortensen, D.L, Tomlinson, E.E, Sadick, M.D, Robinson, I.C, Clark, R.G. | | Deposit date: | 2001-05-11 | | Release date: | 2001-05-30 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of a phage display-derived peptide that binds to insulin-like growth factor binding protein 1.
Biochemistry, 40, 2001
|
|
1GK5
 
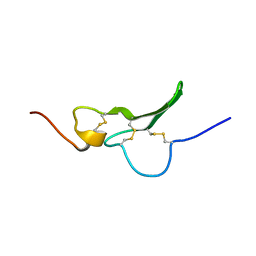 | | Solution Structure the mEGF/TGFalpha44-50 chimeric growth factor | | Descriptor: | Pro-epidermal growth factor,Protransforming growth factor alpha | | Authors: | Chamberlin, S.G, Brennan, L, Puddicombe, S.M, Davies, D.E, Turner, D.L. | | Deposit date: | 2001-08-08 | | Release date: | 2002-08-08 | | Last modified: | 2018-03-28 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Megf/Tgfalpha44-50 Chimeric Growth Factor.
Eur.J.Biochem., 268, 2001
|
|
1G9R
 
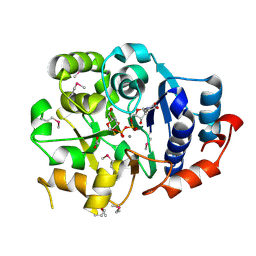 | | CRYSTAL STRUCTURE OF GALACTOSYLTRANSFERASE LGTC IN COMPLEX WITH MN AND UDP-2F-GALACTOSE | | Descriptor: | ACETIC ACID, GLYCOSYL TRANSFERASE, MANGANESE (II) ION, ... | | Authors: | Persson, K, Hoa, D.L, Diekelmann, M, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C.J. | | Deposit date: | 2000-11-27 | | Release date: | 2001-02-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the retaining galactosyltransferase LgtC from Neisseria meningitidis in complex with donor and acceptor sugar analogs.
Nat.Struct.Biol., 8, 2001
|
|
6PD2
 
 | |
6PD1
 
 | |
6F92
 
 | | Structure of the family GH92 alpha-mannosidase BT3965 from Bacteroides thetaiotaomicron in complex with Mannoimidazole (ManI) | | Descriptor: | (5R,6R,7S,8R)-5-(HYDROXYMETHYL)-5,6,7,8-TETRAHYDROIMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Thompson, A.J, Spears, R.J, Zhu, Y, Suits, M.D.L, Williams, S.J, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2017-12-13 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Bacteroides thetaiotaomicron generates diverse alpha-mannosidase activities through subtle evolution of a distal substrate-binding motif.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6F8Z
 
 | | Structure of the family GH92 alpha-mannosidase BT3130 from Bacteroides thetaiotaomicron | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1,2-mannosidase, putative, ... | | Authors: | Thompson, A.J, Spears, R.J, Zhu, Y, Suits, M.D.L, Williams, S.J, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2017-12-13 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Bacteroides thetaiotaomicron generates diverse alpha-mannosidase activities through subtle evolution of a distal substrate-binding motif.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6F91
 
 | | Structure of the family GH92 alpha-mannosidase BT3965 from Bacteroides thetaiotaomicron | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Thompson, A.J, Spears, R.J, Zhu, Y, Suits, M.D.L, Williams, S.J, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2017-12-13 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bacteroides thetaiotaomicron generates diverse alpha-mannosidase activities through subtle evolution of a distal substrate-binding motif.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
9ILB
 
 | | HUMAN INTERLEUKIN-1 BETA | | Descriptor: | PROTEIN (HUMAN INTERLEUKIN-1 BETA) | | Authors: | Yu, B, Blaber, M, Gronenborn, A.M, Clore, G.M, Caspar, D.L.D. | | Deposit date: | 1998-10-22 | | Release date: | 1999-01-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Disordered water within a hydrophobic protein cavity visualized by x-ray crystallography.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
8U45
 
 | | Crystal Structure Analysis of Aspergillus fumigatus alkaline protease | | Descriptor: | Alkaline protease 1, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Fernandez, D, Diec, D.D.L, Guo, W, Russi, S. | | Deposit date: | 2023-09-08 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Targeting Aspergillus allergen oryzin with a chemical probe at atomic precision.
Sci Rep, 13, 2023
|
|
7RSO
 
 | | AMC016 SOSIP.v4.2 in complex with PGV04 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, AMC016 gp120, ... | | Authors: | Bader, D.L.V, Cottrell, C.A, Ward, A.B. | | Deposit date: | 2021-08-11 | | Release date: | 2021-09-15 | | Last modified: | 2022-01-26 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | The Glycan Hole Area of HIV-1 Envelope Trimers Contributes Prominently to the Induction of Autologous Neutralization.
J.Virol., 96, 2022
|
|
5A55
 
 | | The native structure of GH101 from Streptococcus pneumoniae TIGR4 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, ENDO-ALPHA-N-ACETYLGALACTOSAMINIDASE, ... | | Authors: | Gregg, K.J, Suits, M.D.L, Deng, L, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2015-06-16 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Analysis of a Family 101 Glycoside Hydrolase in Complex with Carbohydrates Reveals Insights Into its Mechanism.
J.Biol.Chem., 290, 2015
|
|
5A56
 
 | | The structure of GH101 from Streptococcus pneumoniae TIGR4 in complex with 1-O-methyl-T-antigen | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CITRIC ACID, ... | | Authors: | Gregg, K.J, Suits, M.D.L, Deng, L, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2015-06-16 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Analysis of a Family 101 Glycoside Hydrolase in Complex with Carbohydrates Reveals Insights into Its Mechanism.
J.Biol.Chem., 290, 2015
|
|
5A57
 
 | | The structure of GH101 from Streptococcus pneumoniae TIGR4 in complex with PUGT | | Descriptor: | (Z)-[(3R,4R,5R,6R)-3-acetamido-6-(hydroxymethyl)-4,5-bis(oxidanyl)oxan-2-ylidene]amino] N-phenylcarbamate, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Gregg, K.J, Suits, M.D.L, Deng, L, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2015-06-16 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structural Analysis of a Family 101 Glycoside Hydrolase in Complex with Carbohydrates Reveals Insights Into its Mechanism.
J.Biol.Chem., 290, 2015
|
|
5A59
 
 | | The structure of GH101 E796Q mutant from Streptococcus pneumoniae TIGR4 in complex with T-antigen | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CITRIC ACID, ... | | Authors: | Gregg, K.J, Suits, M.D.L, Deng, L, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2015-06-16 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Analysis of a Family 101 Glycoside Hydrolase in Complex with Carbohydrates Reveals Insights Into its Mechanism.
J.Biol.Chem., 290, 2015
|
|
