6HJN
 
 | | Structure of Influenza Hemagglutinin ectodomain (A/duck/Alberta/35/76) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Benton, D.J, Rosenthal, P.B. | | Deposit date: | 2018-09-04 | | Release date: | 2018-09-26 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Influenza hemagglutinin membrane anchor.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3J48
 
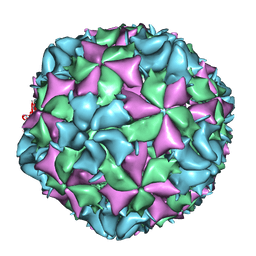 | | Cryo-EM structure of Poliovirus 135S particles | | Descriptor: | Protein VP1, Protein VP2, Protein VP3 | | Authors: | Butan, C, Fiman, D.J, Hogle, J.M. | | Deposit date: | 2013-06-28 | | Release date: | 2013-12-04 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Cryo-Electron Microscopy Reconstruction Shows Poliovirus 135S Particles Poised for Membrane Interaction and RNA Release.
J.Virol., 88, 2014
|
|
6BD1
 
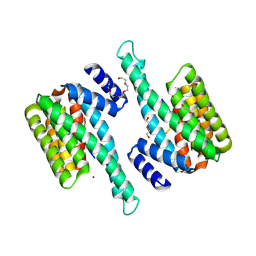 | |
6BCY
 
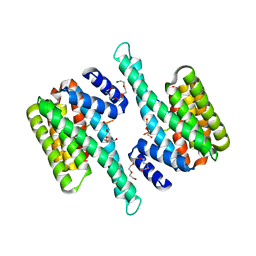 | |
6B48
 
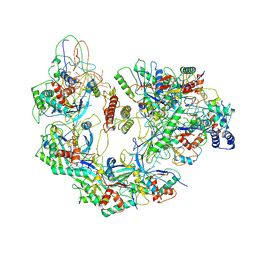 | | Cryo-EM structure of Type I-F CRISPR crRNA-guided Csy surveillance complex with bound anti-CRISPR protein AcrF10 | | Descriptor: | Anti-CRISPR protein AcrF10, CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, ... | | Authors: | Guo, T.W, Bartesaghi, A, Yang, H, Falconieri, V, Rao, P, Merk, A, Fox, T, Earl, L, Patel, D.J, Subramaniam, S. | | Deposit date: | 2017-09-25 | | Release date: | 2017-10-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM Structures Reveal Mechanism and Inhibition of DNA Targeting by a CRISPR-Cas Surveillance Complex.
Cell, 171, 2017
|
|
6B47
 
 | | Cryo-EM structure of Type I-F CRISPR crRNA-guided Csy surveillance complex with bound anti-CRISPR protein AcrF2 | | Descriptor: | Anti-CRISPR protein AcrF2, CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, ... | | Authors: | Guo, T.W, Bartesaghi, A, Yang, H, Falconieri, V, Rao, P, Merk, A, Fox, T, Earl, L, Patel, D.J, Subramaniam, S. | | Deposit date: | 2017-09-25 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM Structures Reveal Mechanism and Inhibition of DNA Targeting by a CRISPR-Cas Surveillance Complex.
Cell, 171, 2017
|
|
6B46
 
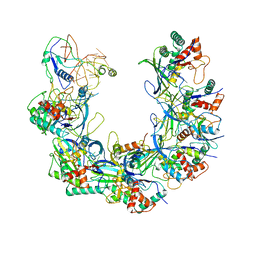 | | Cryo-EM structure of Type I-F CRISPR crRNA-guided Csy surveillance complex with bound anti-CRISPR protein AcrF1 | | Descriptor: | Anti-CRISPR protein AcrF1, CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy3, ... | | Authors: | Guo, T.W, Bartesaghi, A, Yang, H, Falconieri, V, Rao, P, Merk, A, Fox, T, Earl, L, Patel, D.J, Subramaniam, S. | | Deposit date: | 2017-09-25 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM Structures Reveal Mechanism and Inhibition of DNA Targeting by a CRISPR-Cas Surveillance Complex.
Cell, 171, 2017
|
|
6HBW
 
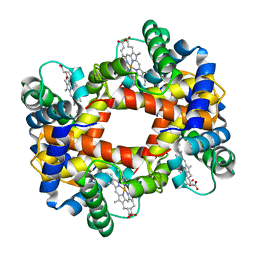 | |
6HJP
 
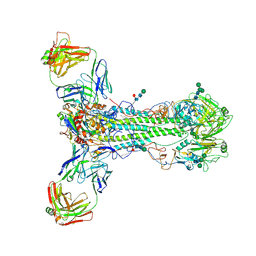 | |
6ASR
 
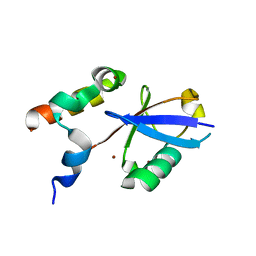 | | REV1 UBM2 domain complex with ubiquitin | | Descriptor: | DNA repair protein REV1, NICKEL (II) ION, Ubiquitin | | Authors: | Miller, D.J. | | Deposit date: | 2017-08-25 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.356 Å) | | Cite: | Structures of REV1 UBM2 Domain Complex with Ubiquitin and with a Small-Molecule that Inhibits the REV1 UBM2-Ubiquitin Interaction.
J. Mol. Biol., 430, 2018
|
|
6AVI
 
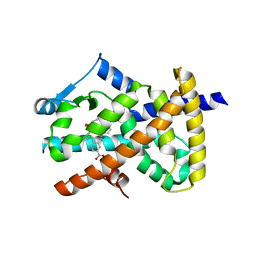 | |
6GYF
 
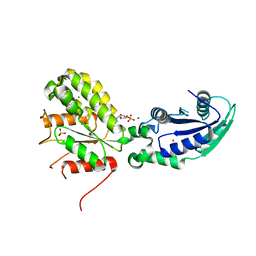 | | Crystal structure of NadR protein in complex with NR | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, MAGNESIUM ION, Nicotinamide-nucleotide adenylyltransferase NadR family / Ribosylnicotinamide kinase, ... | | Authors: | Singh, R, Stetsenko, A, Jaehme, M, Guskov, A, Slotboom, D.J. | | Deposit date: | 2018-06-29 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Functional Characterization of NadR fromLactococcus lactis.
Molecules, 25, 2020
|
|
6GU7
 
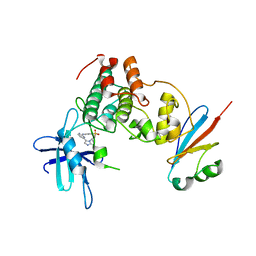 | | CDK1/Cks2 in complex with AZD5438 | | Descriptor: | 4-(2-methyl-3-propan-2-yl-imidazol-4-yl)-~{N}-(4-methylsulfonylphenyl)pyrimidin-2-amine, Cyclin-dependent kinase 1, Cyclin-dependent kinases regulatory subunit 2 | | Authors: | Wood, D.J, Korolchuk, S, Tatum, N.J, Wang, L.Z, Endicott, J.A, Noble, M.E.M, Martin, M.P. | | Deposit date: | 2018-06-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Differences in the Conformational Energy Landscape of CDK1 and CDK2 Suggest a Mechanism for Achieving Selective CDK Inhibition.
Cell Chem Biol, 26, 2019
|
|
1QLK
 
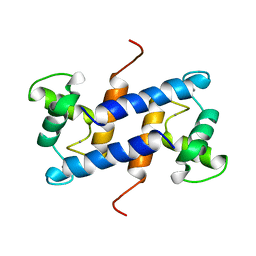 | | SOLUTION STRUCTURE OF CA(2+)-LOADED RAT S100B (BETABETA) NMR, 20 STRUCTURES | | Descriptor: | CALCIUM ION, S-100 PROTEIN | | Authors: | Drohat, A.C, Baldisseri, D.M, Rustandi, R.R, Weber, D.J. | | Deposit date: | 1997-09-26 | | Release date: | 1998-11-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of calcium-bound rat S100B(betabeta) as determined by nuclear magnetic resonance spectroscopy,.
Biochemistry, 37, 1998
|
|
6BJO
 
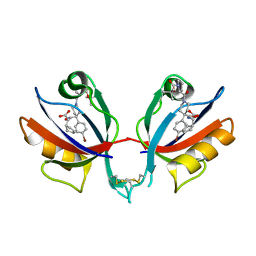 | | PICK1 PDZ domain in complex with the small molecule inhibitor BIO124. | | Descriptor: | (2S)-({4-(4-bromophenyl)-1-[1-(tert-butoxycarbonyl)-L-prolyl]piperidine-4-carbonyl}amino)(cyclopentyl)acetic acid, PRKCA-binding protein | | Authors: | Marcotte, D.J. | | Deposit date: | 2017-11-06 | | Release date: | 2018-01-10 | | Last modified: | 2018-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Lock and chop: A novel method for the generation of a PICK1 PDZ domain and piperidine-based inhibitor co-crystal structure.
Protein Sci., 27, 2018
|
|
6HQN
 
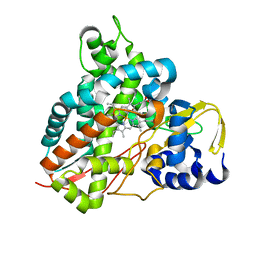 | | Crystal structure of GcoA F169L bound to guaiacol | | Descriptor: | Cytochrome P450, Guaiacol, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mallinson, S.J.B, Hinchen, D.J, Allen, M.D, Johnson, C.W, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2018-09-25 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Enabling microbial syringol conversion through structure-guided protein engineering.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6BJN
 
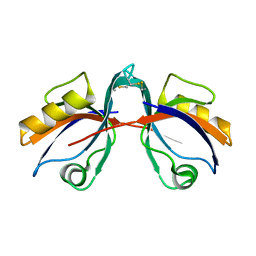 | |
6HQL
 
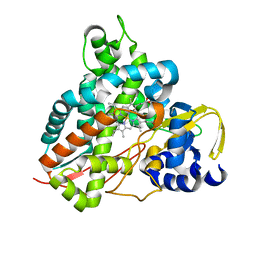 | | Crystal structure of GcoA F169H bound to guaiacol | | Descriptor: | Cytochrome P450, Guaiacol, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mallinson, S.J.B, Hinchen, D.J, Allen, M.D, Johnson, C.W, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2018-09-25 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Enabling microbial syringol conversion through structure-guided protein engineering.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6HQT
 
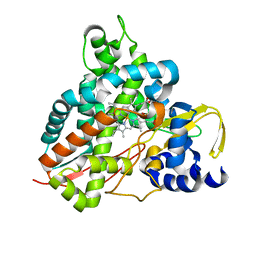 | | Crystal structure of GcoA F169V bound to syringol | | Descriptor: | 2,6-dimethoxyphenol, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mallinson, S.J.B, Hinchen, D.J, Allen, M.D, Johnson, C.W, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2018-09-25 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Enabling microbial syringol conversion through structure-guided protein engineering.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6HS6
 
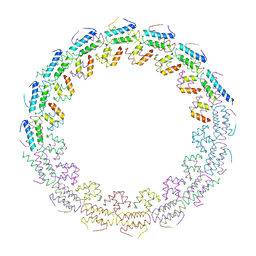 | | C-terminal domain of the TssA component of the type VI secretion system from Burkholderia cenocepacia | | Descriptor: | Type VI secretion protein ImpA | | Authors: | Dix, S.R, Owen, H.J, Sun, R, Ahmad, A, Shastri, S, Spiewak, H.L, Mosby, D.J, Harris, M.J, Batters, S.L, Brooker, T.A, Tzokov, S.B, Sedelnikova, S.E, Baker, P.J, Bullough, P.A, Rice, D.W, Thomas, M.S. | | Deposit date: | 2018-09-28 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Structural insights into the function of type VI secretion system TssA subunits.
Nat Commun, 9, 2018
|
|
6HQO
 
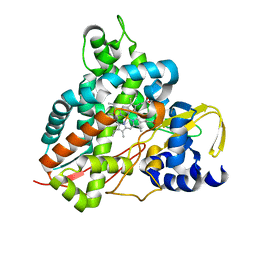 | | Crystal structure of GcoA F169S bound to guaiacol | | Descriptor: | Cytochrome P450, Guaiacol, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mallinson, S.J.B, Hinchen, D.J, Allen, M.D, Johnson, C.W, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2018-09-25 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Enabling microbial syringol conversion through structure-guided protein engineering.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6B89
 
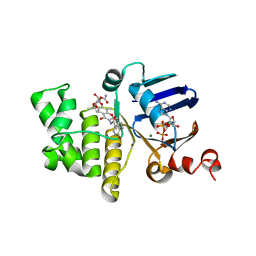 | | E. coli LptB in complex with ADP and novobiocin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lipopolysaccharide export system ATP-binding protein LptB, MAGNESIUM ION, ... | | Authors: | May, J.M, Lazarus, M.B, Sherman, D.J, Owens, T.W, Mandler, M.D, Kahne, D.K. | | Deposit date: | 2017-10-05 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Antibiotic Novobiocin Binds and Activates the ATPase That Powers Lipopolysaccharide Transport.
J. Am. Chem. Soc., 139, 2017
|
|
6I5R
 
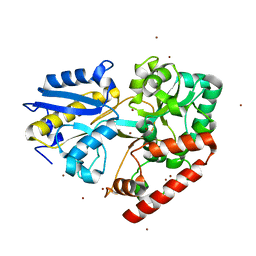 | | BlMnBP1 binding protein of an ABC transporter from Bifidobacterium animalis subsp. lactis ATCC27673 in complex with mannobiose | | Descriptor: | Sugar ABC transporter substrate-binding protein, BlMnBP1, ZINC ION, ... | | Authors: | Abou Hachem, M, Ejby, M, Guskov, A, Slotboom, D.J. | | Deposit date: | 2018-11-14 | | Release date: | 2019-04-17 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Two binding proteins of the ABC transporter that confers growth of Bifidobacterium animalis subsp. lactis ATCC27673 on beta-mannan possess distinct manno-oligosaccharide-binding profiles.
Mol.Microbiol., 112, 2019
|
|
6ANW
 
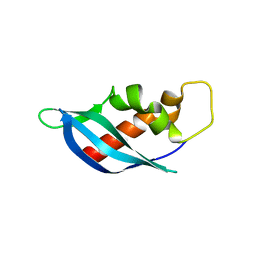 | | Crystal structure of anti-CRISPR protein AcrF10 | | Descriptor: | anti-CRISPR protein AcrF10 | | Authors: | Yang, H, Patel, D.J. | | Deposit date: | 2017-08-14 | | Release date: | 2017-10-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.486 Å) | | Cite: | Cryo-EM Structures Reveal Mechanism and Inhibition of DNA Targeting by a CRISPR-Cas Surveillance Complex.
Cell, 171, 2017
|
|
6HJQ
 
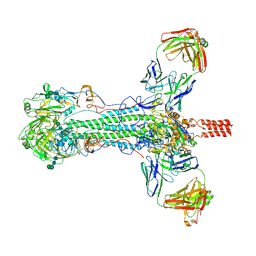 | |
