6BJO
 
 | | PICK1 PDZ domain in complex with the small molecule inhibitor BIO124. | | Descriptor: | (2S)-({4-(4-bromophenyl)-1-[1-(tert-butoxycarbonyl)-L-prolyl]piperidine-4-carbonyl}amino)(cyclopentyl)acetic acid, PRKCA-binding protein | | Authors: | Marcotte, D.J. | | Deposit date: | 2017-11-06 | | Release date: | 2018-01-10 | | Last modified: | 2018-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Lock and chop: A novel method for the generation of a PICK1 PDZ domain and piperidine-based inhibitor co-crystal structure.
Protein Sci., 27, 2018
|
|
6BJN
 
 | |
2NOS
 
 | | MURINE INDUCIBLE NITRIC OXIDE SYNTHASE OXYGENASE DOMAIN (DELTA 114), AMINOGUANIDINE COMPLEX | | Descriptor: | AMINOGUANIDINE, IMIDAZOLE, INDUCIBLE NITRIC OXIDE SYNTHASE, ... | | Authors: | Crane, B.R, Arvai, A.S, Getzoff, E.D, Stuehr, D.J, Tainer, J.A. | | Deposit date: | 1997-09-28 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of nitric oxide synthase oxygenase domain and inhibitor complexes.
Science, 278, 1997
|
|
4CFW
 
 | | Structure-based design of C8-substituted O6-cyclohexylmethoxyguanine CDK1 and 2 inhibitors. | | Descriptor: | 3-[2-amino-6-(cyclohexylmethoxy)-7H-purin-8-yl]-2-methylbenzenesulfonamide, CYCLIN-A2, CYCLIN-DEPENDENT KINASE 2 | | Authors: | Carbain, B, Paterson, D.J, Anscombe, E, Campbell, A, Cano, C, Echalier, A, Endicott, J, Golding, B.T, Haggerty, K, Hardcastle, I.R, Jewsbury, P, Newell, D.R, Noble, M.E.M, Roche, C, Wang, L.Z, Griffin, R. | | Deposit date: | 2013-11-19 | | Release date: | 2013-12-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | 8-Substituted O6-Cyclohexylmethylguanine Cdk2 Inhibitors; Using Structure-Based Inhibitor Design to Optimise an Alternative Binding Mode.
J.Med.Chem., 57, 2014
|
|
5UG1
 
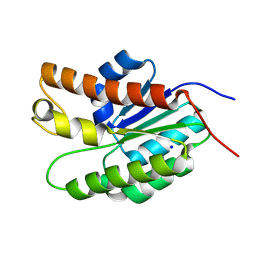 | | Structure of Streptococcus pneumoniae peptidoglycan O-acetyltransferase A (OatA) C-terminal catalytic domain with methylsulfonyl adduct | | Descriptor: | Acyltransferase, SODIUM ION, methanesulfonic acid | | Authors: | Sychantha, D, Jones, C, Little, D.J, Moynihan, P.J, Robinson, H, Galley, N.F, Roper, D.I, Dowson, C.G, Howell, P.L, Clarke, A.J. | | Deposit date: | 2017-01-06 | | Release date: | 2017-10-25 | | Last modified: | 2017-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | In vitro characterization of the antivirulence target of Gram-positive pathogens, peptidoglycan O-acetyltransferase A (OatA).
PLoS Pathog., 13, 2017
|
|
5UFY
 
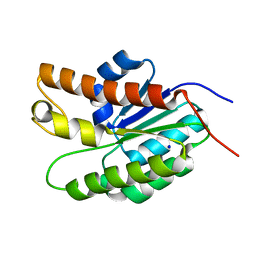 | | Structure of Streptococcus pneumoniae peptidoglycan O-acetyltransferase A (OatA) C-terminal catalytic domain | | Descriptor: | Acyltransferase, SODIUM ION | | Authors: | Sychantha, D, Jones, C, Little, D.J, Moynihan, P.J, Robinson, H, Galley, N.F, Roper, D.I, Dowson, C.G, Howell, P.L, Clarke, A.J. | | Deposit date: | 2017-01-06 | | Release date: | 2017-10-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | In vitro characterization of the antivirulence target of Gram-positive pathogens, peptidoglycan O-acetyltransferase A (OatA).
PLoS Pathog., 13, 2017
|
|
5UGM
 
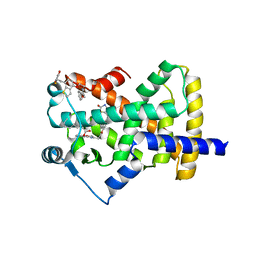 | | Crystal Structure of Human PPARgamma Ligand Binding Domain in Complex with Edaglitazone | | Descriptor: | (5R)-5-({4-[2-(5-methyl-2-phenyl-1,3-oxazol-4-yl)ethoxy]-1-benzothiophen-7-yl}methyl)-1,3-thiazolidine-2,4-dione, Peroxisome proliferator-activated receptor gamma, nonanoic acid | | Authors: | Shang, J, Kojetin, D.J. | | Deposit date: | 2017-01-09 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Cooperative cobinding of synthetic and natural ligands to the nuclear receptor PPAR gamma.
Elife, 7, 2018
|
|
5UHL
 
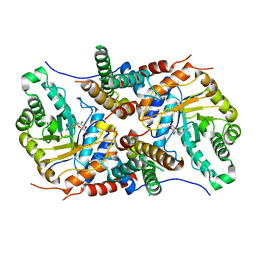 | | Crystal structure of the core catalytic domain of human O-GlcNAcase complexed with Thiamet G | | Descriptor: | (2Z,3aR,5R,6S,7R,7aR)-2-(ethylimino)-5-(hydroxymethyl)hexahydro-3aH-pyrano[3,2-d][1,3]thiazole-6,7-diol, O-GlcNAcase TIM-barrel domain, O-GlcNAcase stalk domain | | Authors: | Klein, D.J, Elsen, N.L. | | Deposit date: | 2017-01-11 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Crystal structure of the core catalytic domain of human O-GlcNAcase and molecular basis of activity and inhibition
To Be Published
|
|
5UHO
 
 | |
4A7N
 
 | | Structure of bare F-actin filaments obtained from the same sample as the Actin-Tropomyosin-Myosin Complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, F-ACTIN | | Authors: | Behrmann, E, Mueller, M, Penczek, P.A, Mannherz, H.G, Manstein, D.J, Raunser, S. | | Deposit date: | 2011-11-14 | | Release date: | 2012-08-01 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Structure of the Rigor Actin-Tropomyosin-Myosin Complex.
Cell(Cambridge,Mass.), 150, 2012
|
|
6BL9
 
 | |
4A3P
 
 | | Structure of USP15 DUSP-UBL deletion mutant | | Descriptor: | ACETATE ION, IODIDE ION, UBIQUITIN CARBOXYL-TERMINAL HYDROLASE 15 | | Authors: | Elliott, P.R, Liu, H, Pastok, M.W, Grossmann, G.J, Rigden, D.J, Clague, M.J, Urbe, S, Barsukov, I.L. | | Deposit date: | 2011-10-03 | | Release date: | 2011-11-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Variability of the Ubiquitin Specific Protease Dusp-Ubl Double Domains.
FEBS Lett., 585, 2011
|
|
5SYS
 
 | | c-Src V281C bound to N-[3-({6-[(1E)-2-cyano-3-(methylamino)-3-oxoprop-1-en-1-yl]-7-(2-methoxyethyl)-7H-pyrrolo[2,3-d]pyrimidin-5-yl}ethynyl)-4-methylphenyl]-3-(trifluoromethyl)benzamide inhibitor | | Descriptor: | N-[3-({6-[(2S)-2-cyano-3-(methylamino)-3-oxopropyl]-7-(2-methoxyethyl)-7H-pyrrolo[2,3-d]pyrimidin-5-yl}ethynyl)-4-methylphenyl]-3-(trifluoromethyl)benzamide, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Dieter, E.M, Merritt, E.A, Maly, D.J. | | Deposit date: | 2016-08-11 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A combined approach reveals a regulatory mechanism coupling Src's kinase activity, localization, and phosphotransferase-independent functions
Mol.Cell, 2019
|
|
6QLW
 
 | | Cathepsin-K in complex with MIV-710 | | Descriptor: | CHLORIDE ION, Cathepsin K, SODIUM ION, ... | | Authors: | Derbyshire, D.J. | | Deposit date: | 2019-02-01 | | Release date: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Successful development of 3-oxohexahydrofuropyrrole amino acid amides as inhibitors of Cathepsin-K.
To Be Published
|
|
5TBX
 
 | | hnRNP A18 RNA Recognition Motif | | Descriptor: | ACETATE ION, Cold-inducible RNA-binding protein, NICKEL (II) ION | | Authors: | Coburn, K.M, Melville, Z, Aligholizadeh, E, Roth, B.M, Varney, K.M, Weber, D.J. | | Deposit date: | 2016-09-13 | | Release date: | 2017-04-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.767 Å) | | Cite: | Crystal structure of the human heterogeneous ribonucleoprotein A18 RNA-recognition motif.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
4CXQ
 
 | | Mycobaterium tuberculosis transaminase BioA complexed with substrate KAPA | | Descriptor: | 1,2-ETHANEDIOL, 7-KETO-8-AMINOPELARGONIC ACID, ADENOSYLMETHIONINE-8-AMINO-7-OXONONANOATE AMINOTRANSFERASE, ... | | Authors: | Dai, R, Wilson, D.J, Geders, T.W, Aldrich, C.C, Finzel, B.C. | | Deposit date: | 2014-04-08 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Inhibition of Mycobacterium Tuberculosis Transaminase Bioa by Aryl Hydrazines and Hydrazides.
Chembiochem, 15, 2014
|
|
5TLU
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the PEG-linked Dimeric Estrogen, EE2-(eg)6-EE2-amine | | Descriptor: | (14beta,17alpha)-21-(4-aminophenyl)-19-norpregna-1(10),2,4-trien-20-yne-3,17-diol, Estrogen receptor, NUCLEAR RECEPTOR COACTIVATOR 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-12 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.223 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5TM3
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with 2,3-bis(2-chloro-4-hydroxyphenyl)thiophene 1-oxide | | Descriptor: | (1S)-2,3-bis(2-chloro-4-hydroxyphenyl)-1H-1lambda~4~-thiophen-1-one, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Erumbi, R, Srinivasan, S, Bruno, N.E, Nowak, J, Izard, T, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-12 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5TML
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the OBHS-ASC compound, (E)-6-(4-((1R,4S,6R)-6-((3-chlorophenoxy)sulfonyl)-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl)phenyl)hex-5-enoic acid | | Descriptor: | 6-{4-[(1S,4S,6R)-6-[(3-chlorophenoxy)sulfonyl]-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl]phenyl}hex-5-enoic acid, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Erumbi, R, Srinivasan, S, Bruno, N.E, Nowak, J, Izard, T, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-13 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5TMV
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the OBHS analog, 4-iodophenyl (1S,2R,4S)-5,6-bis(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonate | | Descriptor: | 4-iodophenyl (1S,2R,4S)-5,6-bis(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonate, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Erumbi, R, Srinivasan, S, Bruno, N.E, Nowak, J, Izard, T, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-13 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5TN5
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the AC-ring estrogen, (1S,3aS,5S,7aS)-5-(4-hydroxyphenyl)-7a-methyloctahydro-1H-inden-1-ol | | Descriptor: | (1S,3aS,5S,7aS)-5-(4-hydroxyphenyl)-7a-methyloctahydro-1H-inden-1-ol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-13 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
6CT4
 
 | | TFE-induced NMR structure of an antimicrobial peptide (EcDBS1R5) derived from a mercury transporter protein (MerP - Escherichia coli) | | Descriptor: | EcDBS1R5 | | Authors: | Cardoso, M.H, Chan, L.Y, Candido, E.S, Craik, D.J, Franco, O.L. | | Deposit date: | 2018-03-22 | | Release date: | 2018-11-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A Computationally Designed Peptide Derived from Escherichia coli as a Potential Drug Template for Antibacterial and Antibiofilm Therapies.
ACS Infect Dis, 4, 2018
|
|
6CG1
 
 | | Crystal Structure of KDM4A with Compound 14 | | Descriptor: | 3-{[(4-fluorophenyl)methyl]amino}pyridine-4-carboxylic acid, Lysine-specific demethylase 4A, NICKEL (II) ION, ... | | Authors: | Hosfield, D.J, Nie, Z. | | Deposit date: | 2018-02-19 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structure-based design and discovery of potent and selective KDM5 inhibitors.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
4CZ2
 
 | | Complex of human VARP-ANKRD1 with Rab32-GppCp. Selenomet derivative. | | Descriptor: | ANKYRIN REPEAT DOMAIN-CONTAINING PROTEIN 27, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, ... | | Authors: | Perez-Dorado, I, Schaefer, I.B, McCoy, A.J, Owen, D.J, Evans, P.R. | | Deposit date: | 2014-04-16 | | Release date: | 2014-06-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Varp is Recruited on to Endosomes by Direct Interaction with Retromer, Where Together They Function in Export to the Cell Surface.
Dev.Cell, 29, 2014
|
|
4AK6
 
 | | BpGH117_H302E mutant glycoside hydrolase | | Descriptor: | ANHYDRO-ALPHA-L-GALACTOSIDASE, MAGNESIUM ION | | Authors: | Hehemann, J.H, Smyth, L, Yadav, A, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2012-02-21 | | Release date: | 2012-03-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Analysis of Keystone Enzyme in Agar Hydrolysis Provides Insight Into the Degradation (of a Polysaccharide from) Red Seaweeds.
J.Biol.Chem., 287, 2012
|
|
