5Y6O
 
 | |
5XZE
 
 | | Mouse cGAS bound to the inhibitor RU332 | | Descriptor: | (3R)-3-[1-(3H-1lambda~4~,3-benzothiazol-2-yl)-5-hydroxy-3-methyl-1H-pyrazol-4-yl]-2-benzofuran-1(3H)-one, Cyclic GMP-AMP synthase, DNA (5'-D(*AP*AP*AP*TP*TP*GP*CP*CP*GP*AP*AP*GP*AP*CP*G)-3'), ... | | Authors: | Vincent, J, Adura, C, Gao, P, Luz, A, Lama, L, Asano, Y, Okamoto, R, Imaeda, T, Aida, J, Rothamel, K, Gogakos, T, Steinberg, J, Reasoner, S, Aso, K, Tuschl, T, Patel, D.J, Glickman, J.F, Ascano, M. | | Deposit date: | 2017-07-12 | | Release date: | 2017-10-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.177 Å) | | Cite: | Small molecule inhibition of cGAS reduces interferon expression in primary macrophages from autoimmune mice.
Nat Commun, 8, 2017
|
|
4W6T
 
 | | Crystal Structure of Full-Length Split GFP Mutant E115H/T118H With Copper Mediated Crystal Contacts, P 43 21 2 Space Group | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, COPPER (II) ION, ... | | Authors: | Leibly, D.J, Waldo, G.S, Yeates, T.O. | | Deposit date: | 2014-08-20 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.604 Å) | | Cite: | A Suite of Engineered GFP Molecules for Oligomeric Scaffolding.
Structure, 23, 2015
|
|
4W76
 
 | | Crystal Structure of Full-Length Split GFP Mutant D21H/K26C Disulfide and Metal-Mediated Dimer, P 21 21 21 Space Group, Form 2 | | Descriptor: | COPPER (II) ION, fluorescent protein D21H/K26C | | Authors: | Leibly, D.J, Waldo, G.S, Yeates, T.O. | | Deposit date: | 2014-08-21 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A Suite of Engineered GFP Molecules for Oligomeric Scaffolding.
Structure, 23, 2015
|
|
4W7E
 
 | | Crystal Structure of Full-Length Split GFP Mutant E124H/K126H With Copper Mediated Crystal Contacts, P 41 21 2 Space Group | | Descriptor: | COPPER (II) ION, IMIDAZOLE, fluorescent protein D21H/K26H | | Authors: | Leibly, D.J, Waldo, G.S, Yeates, T.O. | | Deposit date: | 2014-08-21 | | Release date: | 2015-02-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.592 Å) | | Cite: | A Suite of Engineered GFP Molecules for Oligomeric Scaffolding.
Structure, 23, 2015
|
|
4W7R
 
 | | Crystal Structure of Full-Length Split GFP Mutant E124H/K126H Copper Mediated Dimer, P 21 Space Group | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, fluorescent protein E124H/K126C | | Authors: | Leibly, D.J, Waldo, G.S, Yeates, T.O. | | Deposit date: | 2014-08-22 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Suite of Engineered GFP Molecules for Oligomeric Scaffolding.
Structure, 23, 2015
|
|
4W6N
 
 | |
4W6B
 
 | | Crystal Structure of a Superfolder GFP Mutant K26C Disulfide Dimer, P 21 21 21 Space Group | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, fluorescent protein K26C | | Authors: | Pashkov, I, Sawaya, M.R, Leibly, D.J, Waldo, G.S, Yeates, T.O. | | Deposit date: | 2014-08-20 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Suite of Engineered GFP Molecules for Oligomeric Scaffolding.
Structure, 23, 2015
|
|
4W6G
 
 | |
4W6J
 
 | | Crystal Structure of Full-Length Split GFP Mutant D117C Disulfide Dimer, P 31 2 1 Space Group | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, fluorescent protein D117C | | Authors: | Leibly, D.J, Waldo, G.S, Yeates, T.O. | | Deposit date: | 2014-08-20 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | A Suite of Engineered GFP Molecules for Oligomeric Scaffolding.
Structure, 23, 2015
|
|
4W72
 
 | | Crystal Structure of Full-Length Split GFP Mutant E115C/T118H Disulfide Dimer With Copper Mediated Crystal Contacts, P 21 21 21, Form 1 | | Descriptor: | COPPER (II) ION, fluorescent protein E115C/T118H | | Authors: | Leibly, D.J, Waldo, G.S, Yeates, T.O. | | Deposit date: | 2014-08-21 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Suite of Engineered GFP Molecules for Oligomeric Scaffolding.
Structure, 23, 2015
|
|
4W6O
 
 | |
4W7C
 
 | |
4W6R
 
 | |
4W7X
 
 | |
7TZJ
 
 | | SARS CoV-2 PLpro in complex with inhibitor 3k | | Descriptor: | DIMETHYL SULFOXIDE, N-[(3-fluorophenyl)methyl]-1-[(1R)-1-naphthalen-1-ylethyl]piperidine-4-carboxamide, Papain-like protease, ... | | Authors: | Calleja, D.J, Klemm, T, Lechtenberg, B.C, Kuchel, N.W, Lessene, G, Komander, D. | | Deposit date: | 2022-02-15 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Insights Into Drug Repurposing, as Well as Specificity and Compound Properties of Piperidine-Based SARS-CoV-2 PLpro Inhibitors.
Front Chem, 10, 2022
|
|
4WQ2
 
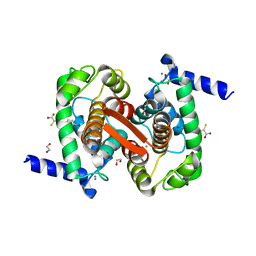 | | Human calpain PEF(S) with (Z)-3-(6-bromondol-3-yl)-2-mercaptoacrylic acid bound | | Descriptor: | (2Z)-3-(6-bromo-1H-indol-3-yl)-2-sulfanylprop-2-enoic acid, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Adams, S.E, Rizkallah, P.J, Miller, D.J, Hallett, M.B, Allemann, R.K. | | Deposit date: | 2014-10-21 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Conformationally restricted calpain inhibitors.
Chem Sci, 6, 2015
|
|
7U7H
 
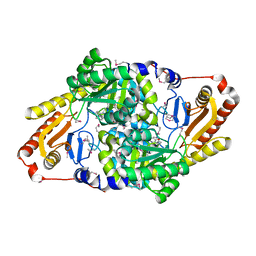 | | Cysteate acyl-ACP transferase from Alistipes finegoldii | | Descriptor: | 7-keto-8-aminopelargonate synthetase-like enzyme, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Radka, C.D, Miller, D.J, Frank, M.W, Rock, C.O. | | Deposit date: | 2022-03-07 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Biochemical characterization of the first step in sulfonolipid biosynthesis in Alistipes finegoldii.
J.Biol.Chem., 298, 2022
|
|
4WNN
 
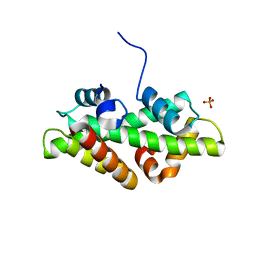 | | SPT16-H2A-H2B FACT HISTONE Complex | | Descriptor: | Histone H2A.1, Histone H2B.1, PHOSPHATE ION, ... | | Authors: | Kemble, D.J, Hill, C.P, Whitby, F.G, Formosa, T, McCullough, L.L. | | Deposit date: | 2014-10-13 | | Release date: | 2015-10-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | FACT Disrupts Nucleosome Structure by Binding H2A-H2B with Conserved Peptide Motifs.
Mol.Cell, 60, 2015
|
|
7UO0
 
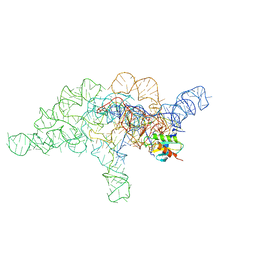 | | E.coli RNaseP Holoenzyme with Mg2+ | | Descriptor: | CALCIUM ION, Precursor tRNA substrate G(-1) G(-2), RNase P RNA, ... | | Authors: | Huang, W, Taylor, D.J. | | Deposit date: | 2022-04-12 | | Release date: | 2022-09-28 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural and mechanistic basis for recognition of alternative tRNA precursor substrates by bacterial ribonuclease P.
Nat Commun, 13, 2022
|
|
7UO1
 
 | | E.coli RNaseP Holoenzyme with Mg2+ | | Descriptor: | CALCIUM ION, E.coli RNase P RNA, Precursor tRNA substrate U(-1) and A(-2), ... | | Authors: | Huang, W, Taylor, D.J. | | Deposit date: | 2022-04-12 | | Release date: | 2022-09-28 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and mechanistic basis for recognition of alternative tRNA precursor substrates by bacterial ribonuclease P.
Nat Commun, 13, 2022
|
|
7UO2
 
 | | E.coli RNaseP Holoenzyme with Mg2+ | | Descriptor: | MAGNESIUM ION, RNase P RNA, Ribonuclease P protein component | | Authors: | Huang, W, Taylor, D.J. | | Deposit date: | 2022-04-12 | | Release date: | 2022-09-28 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and mechanistic basis for recognition of alternative tRNA precursor substrates by bacterial ribonuclease P.
Nat Commun, 13, 2022
|
|
7UO5
 
 | | E.coli RNaseP Holoenzyme with Mg2+ | | Descriptor: | CALCIUM ION, RNase P RNA, Ribonuclease P protein component | | Authors: | Huang, W, Taylor, D.J. | | Deposit date: | 2022-04-12 | | Release date: | 2022-09-28 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and mechanistic basis for recognition of alternative tRNA precursor substrates by bacterial ribonuclease P.
Nat Commun, 13, 2022
|
|
4WQ3
 
 | | Human calpain PEF(S) with (2Z,2Z')-2,2'-disulfanediylbis(3-(6-bromoindol-3-yl)acrylic acid) bound | | Descriptor: | (2Z)-3-(6-bromo-1H-indol-3-yl)-2-sulfanylprop-2-enoic acid, CALCIUM ION, Calpain small subunit 1 | | Authors: | Adams, S.E, Robinson, E.J, Rizkallah, P.J, Miller, D.J, Hallett, M.B, Allemann, R.K. | | Deposit date: | 2014-10-21 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Conformationally restricted calpain inhibitors.
Chem Sci, 6, 2015
|
|
7UDQ
 
 | | Crystal structure of COQ8A-CA157 inhibitor complex in space group P1 | | Descriptor: | 4-[(3,4,5-trimethoxyphenyl)amino]quinoline-7-carbonitrile, Atypical kinase COQ8A, mitochondrial, ... | | Authors: | Bingman, C.A, Murray, N, Smith, R.W, Pagliarini, D.J. | | Deposit date: | 2022-03-20 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Small-molecule inhibition of the archetypal UbiB protein COQ8.
Nat.Chem.Biol., 19, 2023
|
|
