6O0W
 
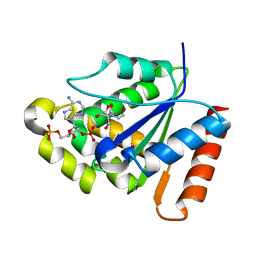 | | Crystal structure of the TIR domain from the grapevine disease resistance protein RUN1 in complex with NADP+ and Bis-Tris | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-2'-5'-DIPHOSPHATE, TIR-NB-LRR type resistance protein RUN1 | | Authors: | Horsefield, S, Burdett, H, Zhang, X, Manik, M.K, Shi, Y, Chen, J, Tiancong, Q, Gilley, J, Lai, J, Gu, W, Rank, M, Casey, L, Ericsson, D.J, Foley, G, Hughes, R.O, Bosanac, T, von Itzstein, M, Rathjen, J.P, Nanson, J.D, Boden, M, Dry, I.B, Williams, S.J, Staskawicz, B.J, Coleman, M.P, Ve, T, Dodds, P.N, Kobe, B. | | Deposit date: | 2019-02-17 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | NAD+cleavage activity by animal and plant TIR domains in cell death pathways.
Science, 365, 2019
|
|
4ZM1
 
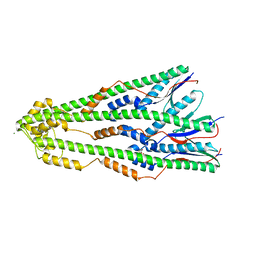 | | Shigella flexneri lipopolysaccharide O-antigen chain-length regulator WzzBSF - wild type | | Descriptor: | CITRIC ACID, Chain length determinant protein, MAGNESIUM ION | | Authors: | Ericsson, D.J, Chang, C.-W, Lonhienne, T, Casey, L, Benning, F, Kobe, B, Tran, E.N.H, Morona, R. | | Deposit date: | 2015-05-02 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural and Biochemical Analysis of a Single Amino-Acid Mutant of WzzBSF That Alters Lipopolysaccharide O-Antigen Chain Length in Shigella flexneri.
Plos One, 10, 2015
|
|
6U24
 
 | | NMR solution structure of triazole bridged SFTI-1 | | Descriptor: | 1-methyl-1H-1,2,3-triazole, GLY-ARG-ALA-THR-LYS-SER-ILE-PRO-PRO-ILE-ALA-PHE-PRO-ASP | | Authors: | White, A.M, Harvey, P.J, Durek, T, Craik, D.J. | | Deposit date: | 2019-08-19 | | Release date: | 2020-07-01 | | Last modified: | 2020-07-15 | | Method: | SOLUTION NMR | | Cite: | Application and Structural Analysis of Triazole-Bridged Disulfide Mimetics in Cyclic Peptides.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
3LMA
 
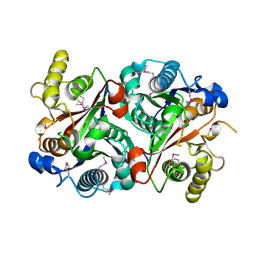 | | Crystal structure of the stage V sporulation protein AD (SpoVAD) from Bacillus licheniformis. Northeast Structural Genomics Consortium Target BiR6. | | Descriptor: | Stage V sporulation protein AD (SpoVAD) | | Authors: | Vorobiev, S, Ashok, S, Seetharaman, J, Belote, R.L, Ciccosanti, C, Patel, D.J, Janjua, H, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-01-29 | | Release date: | 2010-02-09 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (1.993 Å) | | Cite: | Crystal structure of the stage V sporulation protein AD (SpoVAD) from Bacillus licheniformis.
To be Published
|
|
4PE7
 
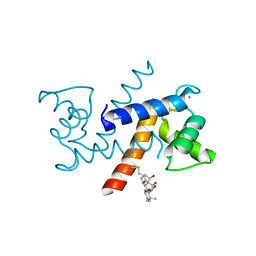 | | Crystal Structure of Calcium-loaded S100B bound to SC1982 | | Descriptor: | (1beta,6beta,7beta,8alpha,9beta,10alpha,13alpha,14R,16beta)-1,6,7,14-tetrahydroxy-7,20-epoxykauran-15-one, CALCIUM ION, Protein S100-B | | Authors: | Cavalier, M.C, Pierce, A.D, Wilder, P.T, Neau, D, Toth, E.A, Weber, D.J. | | Deposit date: | 2014-04-22 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.652 Å) | | Cite: | Covalent Small Molecule Inhibitors of Ca(2+)-Bound S100B.
Biochemistry, 53, 2014
|
|
6O0V
 
 | | Crystal structure of the TIR domain G601P mutant from human SARM1, crystal form 2 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, Sterile alpha and TIR motif-containing protein 1 | | Authors: | Horsefield, S, Burdett, H, Zhang, X, Manik, M.K, Shi, Y, Chen, J, Tiancong, Q, Gilley, J, Lai, J, Gu, W, Rank, M, Casey, L, Ericsson, D.J, Foley, G, Hughes, R.O, Bosanac, T, von Itzstein, M, Rathjen, J.P, Nanson, J.D, Boden, M, Dry, I.B, Williams, S.J, Staskawicz, B.J, Coleman, M.P, Ve, T, Dodds, P.N, Kobe, B. | | Deposit date: | 2019-02-17 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | NAD+cleavage activity by animal and plant TIR domains in cell death pathways.
Science, 365, 2019
|
|
4ZVR
 
 | |
4C1U
 
 | | Structure of the xylo-oligosaccharide specific solute binding protein from Bifidobacterium animalis subsp. lactis Bl-04 in complex with arabinoxylobiose | | Descriptor: | PENTAETHYLENE GLYCOL, SUGAR TRANSPORTER SOLUTE-BINDING PROTEIN, alpha-L-arabinofuranose-(1-3)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Fredslund, F, Ejby, M, Vujicic-Zagar, A, Svensson, B, Slotboom, D.J, Abou Hachem, M. | | Deposit date: | 2013-08-13 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Arabinoxylo-Oligosaccharide Capture by the Probiotic Bifidobacterium Animalis Subsp. Lactis Bl-04
Mol.Microbiol., 90, 2013
|
|
5A55
 
 | | The native structure of GH101 from Streptococcus pneumoniae TIGR4 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, ENDO-ALPHA-N-ACETYLGALACTOSAMINIDASE, ... | | Authors: | Gregg, K.J, Suits, M.D.L, Deng, L, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2015-06-16 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Analysis of a Family 101 Glycoside Hydrolase in Complex with Carbohydrates Reveals Insights Into its Mechanism.
J.Biol.Chem., 290, 2015
|
|
4ZVT
 
 | |
6U7U
 
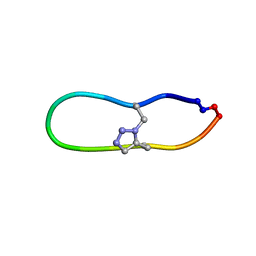 | | NMR solution structure of triazole bridged matriptase inhibitor | | Descriptor: | 1-methyl-1H-1,2,3-triazole, GLY-ARG-ALA-THR-LYS-SER-ILE-PRO-PRO-ARG-ALA-PHE-PRO-ASP | | Authors: | White, A.M, Harvey, P.J, Durek, T, Craik, D.J. | | Deposit date: | 2019-09-03 | | Release date: | 2020-04-22 | | Last modified: | 2020-07-15 | | Method: | SOLUTION NMR | | Cite: | Application and Structural Analysis of Triazole-Bridged Disulfide Mimetics in Cyclic Peptides.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6U9Z
 
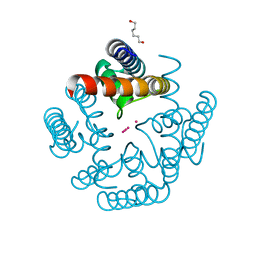 | | Wild-type MthK pore in 6 mM K+ | | Descriptor: | Calcium-gated potassium channel MthK, HEXANE-1,6-DIOL, POTASSIUM ION | | Authors: | Posson, D.J, Nimigean, C.M. | | Deposit date: | 2019-09-09 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.954 Å) | | Cite: | Selectivity filter ion binding affinity determines inactivation in a potassium channel.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4ZVP
 
 | |
5A1J
 
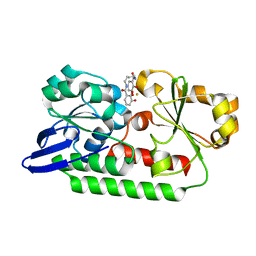 | | Periplasmic Binding Protein CeuE in complex with ferric 4-LICAM | | Descriptor: | ENTEROCHELIN UPTAKE PERIPLASMIC BINDING PROTEIN, FE (III) ION, N,N'-butane-1,4-diylbis(2,3-dihydroxybenzamide) | | Authors: | Raines, D.J, Moroz, O.V, Wilson, K.S, Duhme-Klair, A.K. | | Deposit date: | 2015-04-30 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Interactions of a Periplasmic Binding Protein with a Tetradentate Siderophore Mimic.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
6U9Y
 
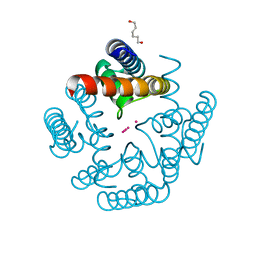 | | Wild-type MthK pore in 11 mM K+ | | Descriptor: | Calcium-gated potassium channel MthK, HEXANE-1,6-DIOL, POTASSIUM ION | | Authors: | Posson, D.J, Nimigean, C.M. | | Deposit date: | 2019-09-09 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Selectivity filter ion binding affinity determines inactivation in a potassium channel.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6U7S
 
 | |
6O0S
 
 | | Crystal structure of the tandem SAM domains from human SARM1 | | Descriptor: | Sterile alpha and TIR motif-containing protein 1 | | Authors: | Horsefield, S, Burdett, H, Zhang, X, Manik, M.K, Shi, Y, Chen, J, Tiancong, Q, Gilley, J, Lai, J, Gu, W, Rank, M, Deerain, N, Casey, L, Ericsson, D.J, Foley, G, Hughes, R.O, Bosanac, T, von Itzstein, M, Rathjen, J.P, Nanson, J.D, Boden, M, Dry, I.B, Williams, S.J, Staskawicz, B.J, Coleman, M.P, Ve, T, Dodds, P.N, Kobe, B. | | Deposit date: | 2019-02-17 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | NAD+cleavage activity by animal and plant TIR domains in cell death pathways.
Science, 365, 2019
|
|
3LD7
 
 | | Crystal structure of the Lin0431 protein from Listeria innocua, Northeast Structural Genomics Consortium Target LkR112 | | Descriptor: | Lin0431 protein | | Authors: | Vorobiev, S, Chen, Y, Lee, D, Patel, D.J, Ciccosanti, C, Sahdev, S, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-01-12 | | Release date: | 2010-01-26 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.547 Å) | | Cite: | Crystal structure of the Lin0431 protein from Listeria innocua
To be Published
|
|
6NXF
 
 | |
6NXL
 
 | | Ubiquitin binding variants | | Descriptor: | Polyubiquitin-B | | Authors: | Miller, D.J, Watson, E.R. | | Deposit date: | 2019-02-08 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Protein engineering of a ubiquitin-variant inhibitor of APC/C identifies a cryptic K48 ubiquitin chain binding site.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
3LLE
 
 | | X-ray structure of bovine SC0322,Ca(2+)-S100B | | Descriptor: | 13-methyl-13,14-dihydro[1,3]benzodioxolo[5,6-c][1,3]dioxolo[4,5-i]phenanthridine, CALCIUM ION, Protein S100-B | | Authors: | Charpentier, T.H, Weber, D.J, Wilder, P.W. | | Deposit date: | 2010-01-28 | | Release date: | 2010-12-29 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | In vitro screening and structural characterization of inhibitors of the S100B-p53 interaction.
Int.J.High Throughput Screen, 2010, 2010
|
|
6U66
 
 | | Structure of the trimeric globular domain of Adiponectin | | Descriptor: | Adiponectin, CALCIUM ION, SODIUM ION | | Authors: | Pascolutti, R, Kruse, A.C, Erlandson, S.C, Burri, D.J, Zheng, S. | | Deposit date: | 2019-08-29 | | Release date: | 2020-01-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Mapping and engineering the interaction between adiponectin and T-cadherin.
J.Biol.Chem., 295, 2020
|
|
6U6N
 
 | | Structure of the trimeric globular domain of Adiponectin mutant - D187A Q188A | | Descriptor: | Adiponectin, CHLORIDE ION | | Authors: | Pascolutti, R, Kruse, A.C, Erlandson, S.C, Burri, D.J, Zheng, S. | | Deposit date: | 2019-08-30 | | Release date: | 2020-01-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Mapping and engineering the interaction between adiponectin and T-cadherin.
J.Biol.Chem., 295, 2020
|
|
6U0K
 
 | | TTBK2 kinase domain in complex with Compound 1 | | Descriptor: | 4-[3-HYDROXYANILINO]-6,7-DIMETHOXYQUINAZOLINE, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Marcotte, D.J, Chodaparambil, J.V. | | Deposit date: | 2019-08-14 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.744 Å) | | Cite: | The crystal structure of the catalytic domain of tau tubulin kinase 2 in complex with a small-molecule inhibitor
Acta Crystallogr.,Sect.F, 76, 2020
|
|
1MFP
 
 | | E. coli Enoyl Reductase in complex with NAD and SB611113 | | Descriptor: | (E)-N-METHYL-N-(1-METHYL-1H-INDOL-3-YLMETHYL)-3-(7-OXO-5,6,7,8-TETRAHYDRO-[1,8]NAPHTHYRIDIN-3-YL)-ACRYLAMIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Seefeld, M.A, Miller, W.H, Newlander, K.A, Burgess, W.J, DeWolf Jr, W.E, Elkins, P.A, Head, M.S, Jakas, D.R, Janson, C.A, Keller, P.M, Manley, P.J, Moore, T.D, Payne, D.J, Pearson, S, Polizzi, B.J, Qiu, X, Rittenhouse, S.F, Uzinskas, I.N, Wallis, N.G, Huffman, W.F. | | Deposit date: | 2002-08-13 | | Release date: | 2003-05-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Indole Naphthyridinones as Inhibitors of Bacterial Enoyl-ACP Reductases FabI and FabK
J.MED.CHEM., 46, 2003
|
|
