1NTB
 
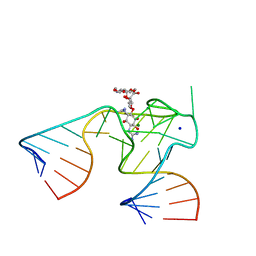 | | 2.9 A crystal structure of Streptomycin RNA-aptamer complex | | 分子名称: | 5'-R(*CP*GP*GP*CP*AP*CP*CP*AP*CP*GP*GP*UP*CP*GP*GP*AP*UP*C)-3', 5'-R(*GP*GP*AP*UP*CP*GP*CP*AP*UP*UP*UP*GP*GP*AP*CP*UP*UP*CP*UP*GP*CP*C)-3', MAGNESIUM ION, ... | | 著者 | Tereshko, V, Skripkin, E, Patel, D.J. | | 登録日 | 2003-01-29 | | 公開日 | 2003-05-13 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Encapsulating Streptomycin within a small 40-mer RNA
CHEM.BIOL., 10, 2003
|
|
2CHM
 
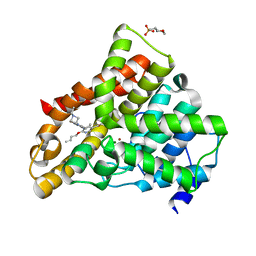 | | Crystal structure of N2 substituted pyrazolo pyrimidinones - a flipped binding mode in PDE5 | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-[2-(BUT-3-EN-1-YLOXY)-5-(1-HYDROXYVINYL)PYRIDIN-3-YL]-3-ETHYL-2-(1-ETHYLAZETIDIN-3-YL)-1,2,6,7A-TETRAHYDRO-7H-PYRAZOLO[4,3-D]PYRIMIDIN-7-ONE, CGMP-SPECIFIC 3', ... | | 著者 | Allerton, C.M.N, Barber, C.G, Beaumont, K.C, Brown, D.G, Cole, S.M, Ellis, D, Lane, C.A.L, Maw, G.N, Mount, N.M, Rawson, D.J, Robinson, C.M, Street, S.D.A, Summerhill, N.W. | | 登録日 | 2006-03-15 | | 公開日 | 2006-06-08 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | A Novel Series of Potent and Selective Pde5 Inhibitors with Potential for High and Dose-Independent Oral Bioavailability
J.Med.Chem., 49, 2006
|
|
1NXG
 
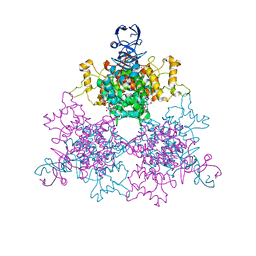 | | The F383A variant of type II Citrate Synthase complexed with NADH | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Citrate synthase, SULFATE ION | | 著者 | Maurus, R, Nguyen, N.T, Stokell, D.J, Ayed, A, Hultin, P.G, Duckworth, H.W, Brayer, G.D. | | 登録日 | 2003-02-10 | | 公開日 | 2003-04-08 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Insights into the evolution of allosteric properties. The NADH binding site of hexameric type II citrate synthases.
Biochemistry, 42, 2003
|
|
2CAY
 
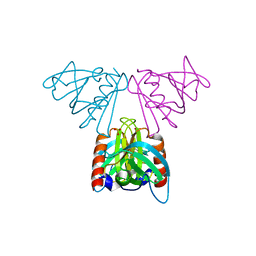 | | Vps36 N-terminal PH domain | | 分子名称: | SULFATE ION, VACUOLAR PROTEIN SORTING PROTEIN 36 | | 著者 | Teo, H, Williams, R.L, Perisic, O, Gill, D.J. | | 登録日 | 2005-12-23 | | 公開日 | 2006-04-07 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Escrt-I Core and Escrt-II Glue Domain Structures Reveal Role for Glue in Linking to Escrt-I and Membranes.
Cell(Cambridge,Mass.), 125, 2006
|
|
5IX1
 
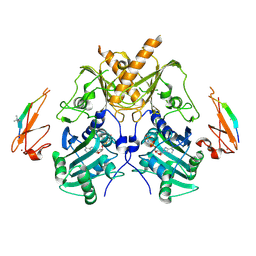 | | Crystal structure of mouse Morc3 ATPase-CW cassette in complex with AMPPNP and H3K4me3 peptide | | 分子名称: | MAGNESIUM ION, MORC family CW-type zinc finger protein 3, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | 著者 | Li, S, Du, J, Patel, D.J. | | 登録日 | 2016-03-23 | | 公開日 | 2016-08-17 | | 最終更新日 | 2016-09-14 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Mouse MORC3 is a GHKL ATPase that localizes to H3K4me3 marked chromatin
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2BAS
 
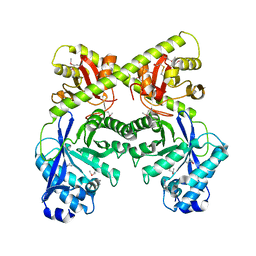 | | Crystal Structure of the Bacillus subtilis YkuI Protein, with an EAL Domain. | | 分子名称: | BETA-MERCAPTOETHANOL, YkuI protein | | 著者 | Minasov, G, Brunzelle, J.S, Shuvalova, L, Miller, D.J, Collart, F.R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2005-10-14 | | 公開日 | 2005-11-29 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.61 Å) | | 主引用文献 | Crystal structures of YkuI and its complex with second messenger cyclic Di-GMP suggest catalytic mechanism of phosphodiester bond cleavage by EAL domains.
J.Biol.Chem., 284, 2009
|
|
5TMQ
 
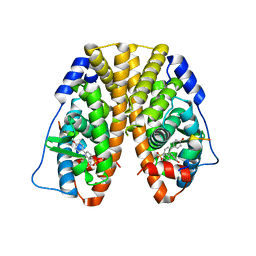 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the Arene Core OBHS derivative, 4-bromophenyl 4,4''-dihydroxy-[1,1':2',1''-terphenyl]-4'-sulfonate | | 分子名称: | 4-bromophenyl 4,4''-dihydroxy-[1,1':2',1''-terphenyl]-4'-sulfonate, Estrogen receptor, Nuclear receptor coactivator 2 | | 著者 | Nwachukwu, J.C, Erumbi, R, Srinivasan, S, Bruno, N.E, Nowak, J, Izard, T, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | 登録日 | 2016-10-13 | | 公開日 | 2017-01-18 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
1O99
 
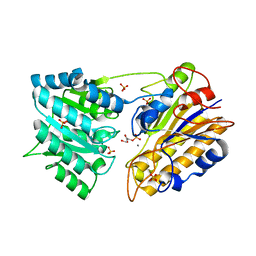 | | CRYSTAL STRUCTURE OF THE S62A MUTANT OF PHOSPHOGLYCERATE MUTASE FROM BACILLUS STEAROTHERMOPHILUS COMPLEXED WITH 2-PHOSPHOGLYCERATE | | 分子名称: | 2,3-BISPHOSPHOGLYCERATE-INDEPENDENT PHOSPHOGLYCERATE MUTASE, 2-PHOSPHOGLYCERIC ACID, MANGANESE (II) ION, ... | | 著者 | Rigden, D.J, Lamani, E, Littlejohn, J.E, Jedrzejas, M.J. | | 登録日 | 2002-12-11 | | 公開日 | 2002-12-23 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Insights Into the Catalytic Mechanism of Cofactor-Independent Phosphoglycerate Mutase from X-Ray Crystallography, Simulated Dynamics and Molecular Modeling
J.Mol.Biol., 328, 2003
|
|
5KX0
 
 | |
5KWP
 
 | |
5KX1
 
 | |
5KVN
 
 | |
2ARG
 
 | | FORMATION OF AN AMINO ACID BINDING POCKET THROUGH ADAPTIVE ZIPPERING-UP OF A LARGE DNA HAIRPIN LOOP, NMR, 9 STRUCTURES | | 分子名称: | ARGININEAMIDE, DNA APTAMER [5'-D (*TP*GP*AP*CP*CP*AP*GP*GP*GP*CP*AP*AP*AP*CP*GP*GP*TP*AP* GP*GP*TP*GP*AP*GP*TP*GP*GP*TP*CP*A)-3'] | | 著者 | Lin, C.H, Wang, W, Jones, R.A, Patel, D.J. | | 登録日 | 1998-08-19 | | 公開日 | 1999-03-23 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Formation of an amino-acid-binding pocket through adaptive zippering-up of a large DNA hairpin loop.
Chem.Biol., 5, 1998
|
|
5TLF
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the Constrained WAY Derivative, 4-(2-(3-methylbut-2-en-1-yl)-7-(trifluoromethyl)-2H-indazol-3-yl)benzene-1,3-diol | | 分子名称: | 4-[2-(3-methylbut-2-en-1-yl)-7-(trifluoromethyl)-2H-indazol-3-yl]benzene-1,3-diol, Estrogen receptor, NUCLEAR RECEPTOR COACTIVATOR 2 | | 著者 | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | 登録日 | 2016-10-11 | | 公開日 | 2017-01-18 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.204 Å) | | 主引用文献 | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
1OB9
 
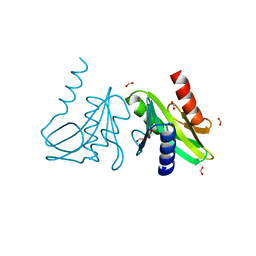 | | Holliday Junction Resolving Enzyme | | 分子名称: | 1,2-ETHANEDIOL, FORMIC ACID, HOLLIDAY JUNCTION RESOLVASE | | 著者 | Middleton, C.L, Parker, J.L, Richard, D.J, White, M.F, Bond, C.S. | | 登録日 | 2003-01-28 | | 公開日 | 2004-10-15 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Substrate Recognition and Catalysis by the Holliday Junction Resolving Enzyme Hje.
Nucleic Acids Res., 32, 2004
|
|
5TLP
 
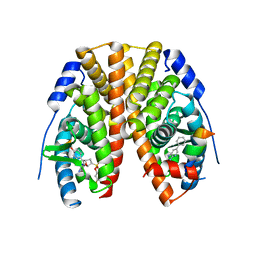 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the OBHS-BSC Analog, 3-fluorophenyl (1R,2R,4S)-5-(4-hydroxyphenyl)-6-(4-(2-(piperidin-1-yl)ethoxy)phenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonate and 3-methyl-6-phenyl-3H-imidazo[4,5-b]pyridin-2-amine | | 分子名称: | 3-fluorophenyl (1S,2R,4S)-5-(4-hydroxyphenyl)-6-{4-[2-(piperidin-1-yl)ethoxy]phenyl}-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonate, 3-methyl-6-phenyl-3H-imidazo[4,5-b]pyridin-2-amine, Estrogen receptor, ... | | 著者 | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | 登録日 | 2016-10-11 | | 公開日 | 2017-01-18 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5TLM
 
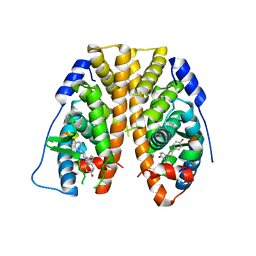 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with 4,4',4''-(thiophene-2,3,5-triyl)triphenol | | 分子名称: | 4,4',4''-(thiene-2,3,5-triyl)triphenol, Estrogen receptor, NUCLEAR RECEPTOR COACTIVATOR 2 | | 著者 | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | 登録日 | 2016-10-11 | | 公開日 | 2017-01-18 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.497 Å) | | 主引用文献 | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5TLY
 
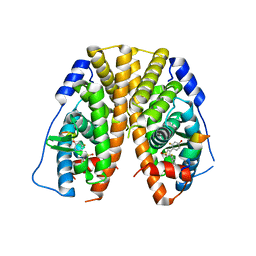 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with 3,4-bis(2-fluoro-4-hydroxyphenyl)thiophene 1,1-dioxide | | 分子名称: | 3,4-bis(2-fluoro-4-hydroxyphenyl)-1H-1lambda~6~-thiophene-1,1-dione, Estrogen receptor, Nuclear receptor coactivator 2 | | 著者 | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | 登録日 | 2016-10-12 | | 公開日 | 2017-01-18 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.143 Å) | | 主引用文献 | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
1ODT
 
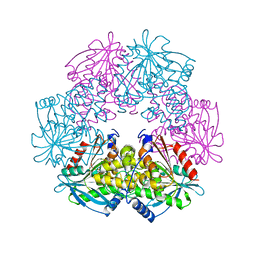 | | cephalosporin C deacetylase mutated, in complex with acetate | | 分子名称: | ACETATE ION, CEPHALOSPORIN C DEACETYLASE | | 著者 | Vincent, F, Charnock, S.J, Verschueren, K.H.G, Turkenburg, J.P, Scott, D.J, Offen, W.A, Roberts, S, Pell, G, Gilbert, H.J, Brannigan, J.A, Davies, G.J. | | 登録日 | 2003-02-20 | | 公開日 | 2003-07-10 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Multifunctional Xylooligosaccharide/Cephalosporin C Deacetylase Revealed by the Hexameric Structure of the Bacillus Subtilis Enzyme at 1.9A Resolution
J.Mol.Biol., 330, 2003
|
|
5TM7
 
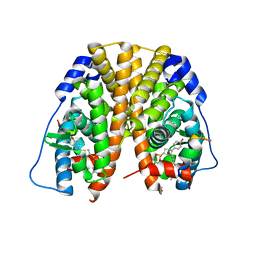 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the OBHS-ASC compound, 7-(4-((1R,4S,6R)-6-((3-chlorophenoxy)sulfonyl)-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl)phenoxy)heptanoic acid | | 分子名称: | 7-{4-[(1S,4S,6R)-6-[(3-chlorophenoxy)sulfonyl]-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl]phenoxy}heptanoic acid, Estrogen receptor, Nuclear receptor coactivator 2 | | 著者 | Nwachukwu, J.C, Erumbi, R, Srinivasan, S, Bruno, N.E, Nowak, J, Izard, T, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | 登録日 | 2016-10-12 | | 公開日 | 2017-01-18 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
1O98
 
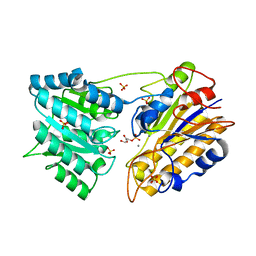 | | 1.4A CRYSTAL STRUCTURE OF PHOSPHOGLYCERATE MUTASE FROM BACILLUS STEAROTHERMOPHILUS COMPLEXED WITH 2-PHOSPHOGLYCERATE | | 分子名称: | 2,3-BISPHOSPHOGLYCERATE-INDEPENDENT PHOSPHOGLYCERATE MUTASE, 2-PHOSPHOGLYCERIC ACID, MANGANESE (II) ION, ... | | 著者 | Rigden, D.J, Lamani, E, Littlejohn, J.E, Jedrzejas, M.J. | | 登録日 | 2002-12-11 | | 公開日 | 2003-05-01 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Insights Into the Catalytic Mechanism of Cofactor-Independent Phosphoglycerate Mutase from X-Ray Crystallography, Simulated Dynamics and Molecular Modeling
J.Mol.Biol., 328, 2003
|
|
5TLV
 
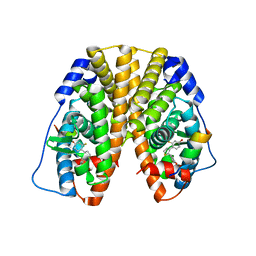 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with 4,4'-(thiophene-2,3-diyl)bis(3-fluorophenol) | | 分子名称: | 4,4'-(thiene-2,3-diyl)bis(3-fluorophenol), Estrogen receptor, Nuclear receptor coactivator 2 | | 著者 | Nwachukwu, J.C, Erumbi, R, Srinivasan, S, Bruno, N.E, Nowak, J, Izard, T, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | 登録日 | 2016-10-12 | | 公開日 | 2017-01-18 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.323 Å) | | 主引用文献 | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5TM4
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the OBHS-ASC Analog, 5-(4-((1R,4S,6R)-6-((3-chlorophenoxy)sulfonyl)-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl)phenoxy)pentanoic acid | | 分子名称: | 5-{4-[(1S,4S,6R)-6-[(3-chlorophenoxy)sulfonyl]-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl]phenoxy}pentanoic acid, Estrogen receptor, Nuclear receptor coactivator 2 | | 著者 | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | 登録日 | 2016-10-12 | | 公開日 | 2017-01-18 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5TMW
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the OBHS derivative, 4-acetamidophenyl (1S,2R,4S)-5,6-bis(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonate | | 分子名称: | 4-(acetylamino)phenyl (1S,2R,4S)-5,6-bis(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonate, Estrogen receptor, Nuclear receptor coactivator 2 | | 著者 | Nwachukwu, J.C, Erumbi, R, Srinivasan, S, Bruno, N.E, Nowak, J, Izard, T, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | 登録日 | 2016-10-13 | | 公開日 | 2017-01-18 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.286 Å) | | 主引用文献 | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5TN3
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the estradiol derivative, (8S,9S,13S,14S)-17-((4-isopropylphenyl)amino)-13-methyl-7,8,9,11,12,13,14,15,16,17-decahydro-6H-cyclopenta[a]phenanthren-3-ol | | 分子名称: | (9beta,13alpha,17beta)-17-{[4-(propan-2-yl)phenyl]amino}estra-1(10),2,4-trien-3-ol, Estrogen receptor, Nuclear receptor coactivator 2 | | 著者 | Nwachukwu, J.C, Erumbi, R, Srinivasan, S, Bruno, N.E, Nowak, J, Izard, T, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | 登録日 | 2016-10-13 | | 公開日 | 2017-01-18 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.543 Å) | | 主引用文献 | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
