7PUA
 
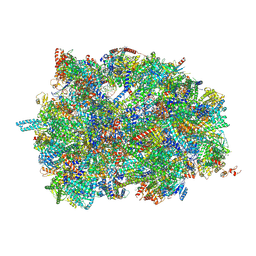 | | Middle assembly intermediate of the Trypanosoma brucei mitoribosomal small subunit | | 分子名称: | 30S Ribosomal protein S17, putative, 30S ribosomal protein S8, ... | | 著者 | Lenarcic, T, Leibundgut, M, Saurer, M, Ramrath, D.J.F, Fluegel, T, Boehringer, D, Ban, N. | | 登録日 | 2021-09-29 | | 公開日 | 2022-03-02 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Mitoribosomal small subunit maturation involves formation of initiation-like complexes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4ONJ
 
 | |
6J23
 
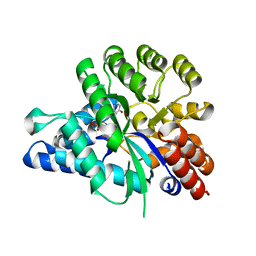 | | Crystal structure of arabidopsis ADAL complexed with GMP | | 分子名称: | Adenosine/AMP deaminase family protein, GUANOSINE-5'-MONOPHOSPHATE, ZINC ION | | 著者 | Wu, B.X, Zhang, D, Nie, H.B, Shen, S.L, Li, S.S, Patel, D.J. | | 登録日 | 2018-12-30 | | 公開日 | 2019-02-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure ofArabidopsis thaliana N6-methyl-AMP deaminase ADAL with bound GMP and IMP and implications forN6-methyl-AMP recognition and processing.
Rna Biol., 16, 2019
|
|
6F24
 
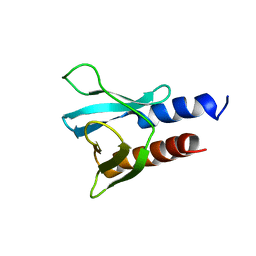 | | PH domain from PfAPH | | 分子名称: | C-terminal PH domain from PfAPH | | 著者 | Darvill, N, Liu, B, Matthews, S, Soldati-Favre, D, Rouse, S, Benjamin, S, Blake, T, Dubois, D.J, Hammoudi, P.M, Pino, P. | | 登録日 | 2017-11-23 | | 公開日 | 2018-12-12 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | C-terminal PH domain from P. falciparum acylated plekstrin homology domain containing protein (APH)
To Be Published
|
|
4ONQ
 
 | |
6J4T
 
 | | Crystal structure of arabidopsis ADAL complexed with IMP | | 分子名称: | Adenosine/AMP deaminase family protein, INOSINIC ACID, ZINC ION | | 著者 | Wu, B.X, Zhang, D, Nie, H.B, Shen, S.L, Li, S.S, Patel, D.J. | | 登録日 | 2019-01-10 | | 公開日 | 2019-07-31 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Structure ofArabidopsis thaliana N6-methyl-AMP deaminase ADAL with bound GMP and IMP and implications forN6-methyl-AMP recognition and processing.
Rna Biol., 16, 2019
|
|
6JFV
 
 | | The crystal structure of 2B-2B complex from keratins 5 and 14 (C367A mutant of K14) | | 分子名称: | Keratin, type I cytoskeletal 14, type II cytoskeletal 5 | | 著者 | Kim, M.S, Lee, C.H, Coulombe, P.A, Leahy, D.J. | | 登録日 | 2019-02-12 | | 公開日 | 2020-01-22 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structure-Function Analyses of a Keratin Heterotypic Complex Identify Specific Keratin Regions Involved in Intermediate Filament Assembly.
Structure, 28, 2020
|
|
6F36
 
 | | Polytomella Fo model | | 分子名称: | Mitochondrial ATP synthase subunit 6, Mitochondrial ATP synthase subunit ASA6, Mitochondrial ATP synthase subunit c | | 著者 | Yildiz, O, Kuehlbrandt, W, Klusch, N, Murphy, B.J, Mills, D.J. | | 登録日 | 2017-11-28 | | 公開日 | 2017-12-20 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural basis of proton translocation and force generation in mitochondrial ATP synthase.
Elife, 6, 2017
|
|
7AM3
 
 | |
7AM6
 
 | |
7AM8
 
 | |
7AM4
 
 | |
7AM5
 
 | |
7AM7
 
 | |
4OUD
 
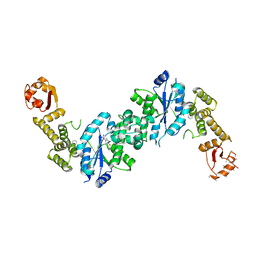 | | Engineered tyrosyl-tRNA synthetase with the nonstandard amino acid L-4,4-biphenylalanine | | 分子名称: | TYROSINE, Tyrosyl-tRNA synthetase | | 著者 | Takeuchi, R, Mandell, D.J, Lajoie, M.J, Church, G.M, Stoddard, B.L. | | 登録日 | 2014-02-16 | | 公開日 | 2015-01-28 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Biocontainment of genetically modified organisms by synthetic protein design.
Nature, 518, 2015
|
|
4OEC
 
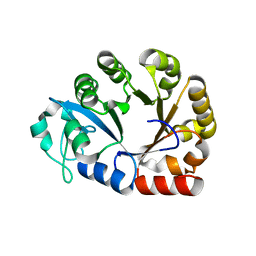 | | Crystal structure of glycerophosphodiester phosphodiesterase from Thermococcus kodakarensis KOD1 | | 分子名称: | Glycerophosphoryl diester phosphodiesterase, MAGNESIUM ION | | 著者 | Atsuta, Y, You, D.J, Takano, K, Koga, Y, Kanaya, S. | | 登録日 | 2014-01-13 | | 公開日 | 2015-01-14 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of glycerophosphodiester phosphodiesterase from Thermococcus kodakarensis KOD1
To be Published
|
|
6F8E
 
 | | PH domain from TgAPH | | 分子名称: | Pleckstrin homology domain | | 著者 | Darvill, N, Liu, B, Matthews, S, Soldati-Favre, D, Rouse, S, Benjamin, S, Blake, T, Dubois, D.J, Hammoudi, P.M, Pino, P. | | 登録日 | 2017-12-13 | | 公開日 | 2019-01-30 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Basis of Phosphatidic Acid Sensing by APH in Apicomplexan Parasites.
Structure, 26, 2018
|
|
4P32
 
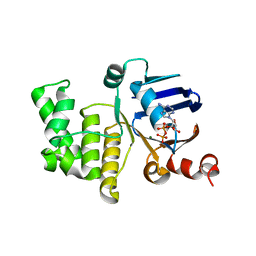 | | Crystal structure of E. coli LptB in complex with ADP-magnesium | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Lipopolysaccharide export system ATP-binding protein LptB, MAGNESIUM ION | | 著者 | Sherman, D.J, Lazarus, M.B, Murphy, L, Liu, C, Walker, S, Ruiz, N, Kahne, D. | | 登録日 | 2014-03-05 | | 公開日 | 2014-03-26 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Decoupling catalytic activity from biological function of the ATPase that powers lipopolysaccharide transport.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4PGF
 
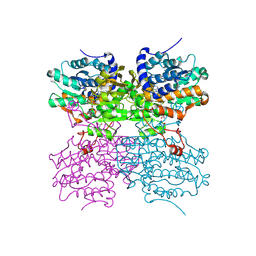 | | The structure of mono-acetylated SAHH | | 分子名称: | ADENOSINE, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Kavran, J.M, Wang, Y, Cole, P.A, Leahy, D.J. | | 登録日 | 2014-05-01 | | 公開日 | 2014-10-01 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.59 Å) | | 主引用文献 | Regulation of s-adenosylhomocysteine hydrolase by lysine acetylation.
J.Biol.Chem., 289, 2014
|
|
4P7L
 
 | | Structure of Escherichia coli PgaB C-terminal domain, P212121 crystal form | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, Poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase | | 著者 | Little, D.J, Li, G, Ing, C, DiFrancesco, B, Bamford, N.C, Robinson, H, Nitz, M, Pomes, R, Howell, P.L. | | 登録日 | 2014-03-27 | | 公開日 | 2014-07-02 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.802 Å) | | 主引用文献 | Modification and periplasmic translocation of the biofilm exopolysaccharide poly-beta-1,6-N-acetyl-D-glucosamine.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5CNQ
 
 | | Crystal structure of the Holliday junction-resolving enzyme GEN1 (WT) in complex with product DNA, Mg2+ and Mn2+ ions | | 分子名称: | DNA (5'-D(*TP*GP*AP*GP*CP*GP*GP*TP*GP*GP*TP*TP*GP*GP*T)-3'), MANGANESE (II) ION, Nuclease-like protein, ... | | 著者 | Liu, Y.J, Freeman, A.D.J, Declais, A.C, Wilson, T.J, Gartner, A, Lilley, D.M.J. | | 登録日 | 2015-07-17 | | 公開日 | 2015-12-30 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.602 Å) | | 主引用文献 | Crystal Structure of a Eukaryotic GEN1 Resolving Enzyme Bound to DNA.
Cell Rep, 13, 2015
|
|
6FKH
 
 | | Chloroplast F1Fo conformation 2 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase delta chain, ... | | 著者 | Hahn, A, Vonck, J, Mills, D.J, Meier, T, Kuehlbrandt, W. | | 登録日 | 2018-01-24 | | 公開日 | 2018-05-23 | | 最終更新日 | 2019-12-11 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Structure, mechanism, and regulation of the chloroplast ATP synthase.
Science, 360, 2018
|
|
6HIX
 
 | | Cryo-EM structure of the Trypanosoma brucei mitochondrial ribosome - This entry contains the large mitoribosomal subunit | | 分子名称: | 12S rRNA, 50S ribosomal protein L13, putative, ... | | 著者 | Ramrath, D.J.F, Niemann, M, Leibundgut, M, Bieri, P, Prange, C, Horn, K, Leitner, A, Boehringer, D, Schneider, A, Ban, N. | | 登録日 | 2018-08-31 | | 公開日 | 2018-09-26 | | 最終更新日 | 2019-02-06 | | 実験手法 | ELECTRON MICROSCOPY (3.39 Å) | | 主引用文献 | Evolutionary shift toward protein-based architecture in trypanosomal mitochondrial ribosomes.
Science, 362, 2018
|
|
4PEF
 
 | | Dbr1 in complex with sulfate | | 分子名称: | GLYCEROL, MANGANESE (II) ION, RNA lariat debranching enzyme, ... | | 著者 | Montemayor, E.J, Katolik, A, Clark, N.E, Taylor, A.B, Schuermann, J.P, Combs, D.J, Johnsson, R, Holloway, S.P, Stevens, S.W, Damha, M.J, Hart, P.J. | | 登録日 | 2014-04-23 | | 公開日 | 2014-08-27 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Structural basis of lariat RNA recognition by the intron debranching enzyme Dbr1.
Nucleic Acids Res., 42, 2014
|
|
7BNO
 
 | | Open conformation of D614G SARS-CoV-2 spike with 2 Erect RBDs | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Benton, D.J, Wrobel, A.G, Rosenthal, P.B, Gamblin, S.J. | | 登録日 | 2021-01-22 | | 公開日 | 2021-02-03 | | 最終更新日 | 2021-03-10 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | The effect of the D614G substitution on the structure of the spike glycoprotein of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
