3IRC
 
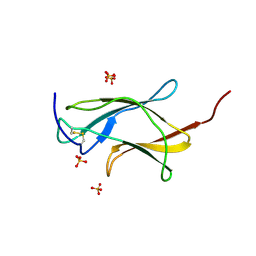 | | Crystal structure analysis of dengue-1 envelope protein domain III | | Descriptor: | ENVELOPE PROTEIN, SULFATE ION | | Authors: | Nelson, C.A, Kim, T, Warren, J.T, Chruszcz, M, Minor, W, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-08-21 | | Release date: | 2009-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure Analysis of the Dengue-1 Envelope Protein Domain III
To be Published
|
|
4QDT
 
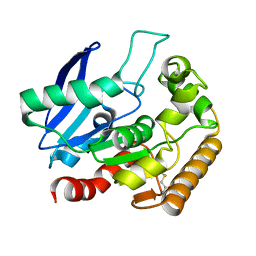 | |
2HFK
 
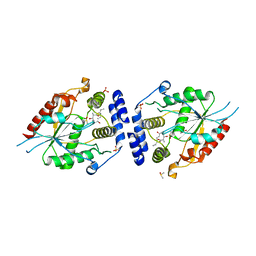 | | Pikromycin thioesterase in complex with product 10-deoxymethynolide | | Descriptor: | (3R,4S,5S,7R,9E,11R,12R)-12-ETHYL-4-HYDROXY-3,5,7,11-TETRAMETHYLOXACYCLODODEC-9-ENE-2,8-DIONE, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Akey, D.L, Kittendorf, J.D, Giraldes, J.W, Fecik, R.A, Sherman, D.H, Smith, J.L. | | Deposit date: | 2006-06-24 | | Release date: | 2006-09-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural Basis for Macrolactonization by the Pikromycin Thioesterase
NAT.CHEM.BIOL., 2, 2006
|
|
2INZ
 
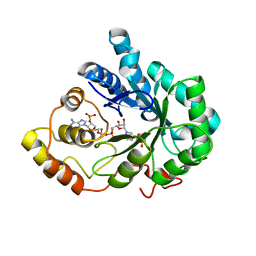 | | Crystal Structure of Aldose Reductase complexed with 2-Hydroxyphenylacetic Acid | | Descriptor: | (2-HYDROXYPHENYL)ACETIC ACID, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Harrison, D.H.T, Pape, E, Carlson, E, Brownlee, J.M. | | Deposit date: | 2006-10-09 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and thermodynamic studies of simple aldose reductase-inhibitor complexes.
Bioorg.Chem., 34, 2006
|
|
2J7O
 
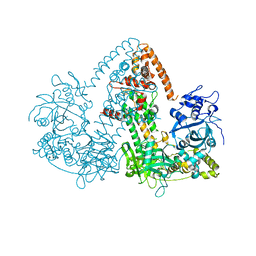 | | STRUCTURE OF THE RNAI POLYMERASE FROM NEUROSPORA CRASSA | | Descriptor: | MAGNESIUM ION, RNA DEPENDENT RNA POLYMERASE | | Authors: | Salgado, P.S, Koivunen, M.R.L, Makeyev, E.V, Bamford, D.H, Stuart, D.I, Grimes, J.M. | | Deposit date: | 2006-10-13 | | Release date: | 2006-12-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The Structure of an Rnai Polymerase Links RNA Silencing and Transcription.
Plos Biol., 4, 2006
|
|
3J7L
 
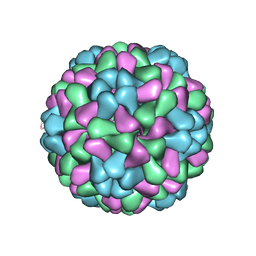 | | Full virus map of brome mosaic virus | | Descriptor: | Capsid protein | | Authors: | Wang, Z, Hryc, C, Bammes, B, Afonine, P.V, Jakana, J, Chen, D.H, Liu, X, Baker, M.L, Kao, C, Ludtke, S.J, Schmid, M.F, Adams, P.D, Chiu, W. | | Deposit date: | 2014-07-18 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | An atomic model of brome mosaic virus using direct electron detection and real-space optimization.
Nat Commun, 5, 2014
|
|
3J7N
 
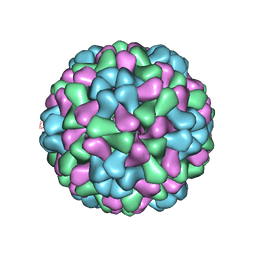 | | Virus model of brome mosaic virus (second half data set) | | Descriptor: | Capsid protein | | Authors: | Wang, Z, Hryc, C, Bammes, B, Afonine, P.V, Jakana, J, Chen, D.H, Liu, X, Baker, M.L, Kao, C, Ludtke, S.J, Schmid, M.F, Adams, P.D, Chiu, W. | | Deposit date: | 2014-07-18 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | An atomic model of brome mosaic virus using direct electron detection and real-space optimization.
Nat Commun, 5, 2014
|
|
2INE
 
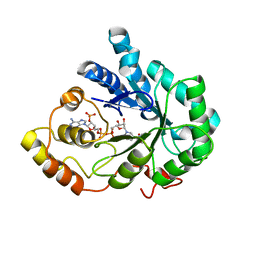 | |
2IS7
 
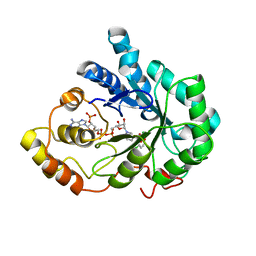 | | Crystal Structure of Aldose Reductase complexed with Dichlorophenylacetic acid | | Descriptor: | (2,6-DICHLOROPHENYL)ACETIC ACID, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Harrison, D.H.T, Milne, A, Brownlee, J.M. | | Deposit date: | 2006-10-16 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and thermodynamic studies of simple aldose reductase-inhibitor complexes.
Bioorg.Chem., 34, 2006
|
|
2IPW
 
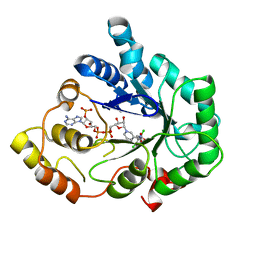 | | Crystal Structure of C298A W219Y Aldose Reductase complexed with Dichlorophenylacetic acid | | Descriptor: | (2,6-DICHLOROPHENYL)ACETIC ACID, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Harrison, D.H.T, Pape, E, Brownlee, J.M. | | Deposit date: | 2006-10-12 | | Release date: | 2006-11-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and thermodynamic studies of simple aldose reductase-inhibitor complexes.
Bioorg.Chem., 34, 2006
|
|
2IQD
 
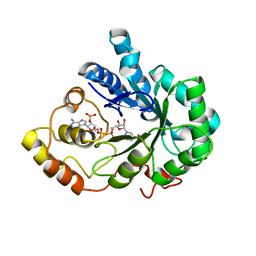 | | Crystal Structure of Aldose Reductase complexed with Lipoic Acid | | Descriptor: | Aldose reductase, LIPOIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Harrison, D.H.T, Carlson, E, Pape, E, Brownlee, J.M. | | Deposit date: | 2006-10-13 | | Release date: | 2006-11-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and thermodynamic studies of simple aldose reductase-inhibitor complexes.
Bioorg.Chem., 34, 2006
|
|
2ISF
 
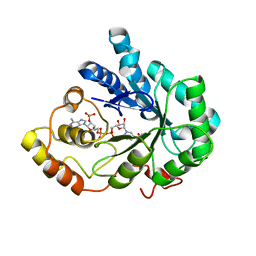 | |
2IQ0
 
 | |
6B19
 
 | | Architecture of HIV-1 reverse transcriptase initiation complex core | | Descriptor: | RNA genome fragment, reverse transcriptase p51 subunit, reverse transcriptase p66 subunit, ... | | Authors: | Larsen, K.P, Mathiharan, Y.K, Chen, D.H, Puglisi, J.D, Skiniotis, G, Puglisi, E.V. | | Deposit date: | 2017-09-18 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Architecture of an HIV-1 reverse transcriptase initiation complex.
Nature, 557, 2018
|
|
2YM7
 
 | | Crystal structure of checkpoint kinase 1 (Chk1) in complex with inhibitors | | Descriptor: | 1,2-ETHANEDIOL, 5-({6-[(piperidin-4-ylmethyl)amino]pyrimidin-4-yl}amino)pyrazine-2-carbonitrile, SERINE/THREONINE-PROTEIN KINASE CHK1 | | Authors: | Reader, J.C, Matthews, T.P, Klair, S, Cheung, K.M.J, Scanlon, J, Proisy, N, Addison, G, Ellard, J, Piton, N, Taylor, S, Cherry, M, Fisher, M, Boxall, K, Burns, S, Walton, M.I, Westwood, I.M, Hayes, A, Eve, P, Valenti, M, Brandon, A.H, Box, G, vanMontfort, R.L.M, Williams, D.H, Aherne, G.W, Raynaud, F.I, Eccles, S.A, Garrett, M.D, Collins, I. | | Deposit date: | 2011-06-06 | | Release date: | 2012-01-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure-Guided Evolution of Potent and Selective Chk1 Inhibitors Through Scaffold Morphing.
J.Med.Chem., 54, 2011
|
|
6B4H
 
 | | Crystal structure of Chaetomium thermophilum Gle1 CTD-Nup42 GBM-IP6 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, INOSITOL HEXAKISPHOSPHATE, Nucleoporin AMO1, ... | | Authors: | Lin, D.H, Correia, A.R, Cai, S.W, Huber, F.M, Jette, C.A, Hoelz, A. | | Deposit date: | 2017-09-26 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural and functional analysis of mRNA export regulation by the nuclear pore complex.
Nat Commun, 9, 2018
|
|
6B4E
 
 | | Crystal structure of Saccharomyces cerevisiae Gle1 CTD-Nup42 GBM complex | | Descriptor: | 1,2-ETHANEDIOL, Nucleoporin GLE1, Nucleoporin NUP42, ... | | Authors: | Lin, D.H, Correia, A.R, Cai, S.W, Huber, F.M, Jette, C.A, Hoelz, A. | | Deposit date: | 2017-09-26 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and functional analysis of mRNA export regulation by the nuclear pore complex.
Nat Commun, 9, 2018
|
|
6B4J
 
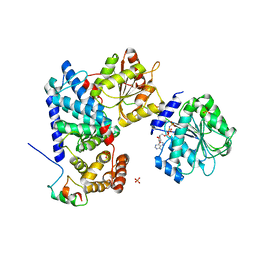 | | Crystal structure of human Gle1 CTD-Nup42 GBM-DDX19B(AMPPNP) complex | | Descriptor: | ATP-dependent RNA helicase DDX19B, MAGNESIUM ION, Nucleoporin GLE1, ... | | Authors: | Lin, D.H, Correia, A.R, Cai, S.W, Huber, F.M, Jette, C.A, Hoelz, A. | | Deposit date: | 2017-09-26 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural and functional analysis of mRNA export regulation by the nuclear pore complex.
Nat Commun, 9, 2018
|
|
6B6N
 
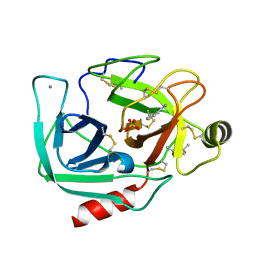 | | Orthorhombic trypsin (295 K) in the presence of 50% mpd | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, BENZAMIDINE, CALCIUM ION, ... | | Authors: | Juers, D.H. | | Deposit date: | 2017-10-03 | | Release date: | 2017-11-01 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The impact of cryosolution thermal contraction on proteins and protein crystals: volumes, conformation and order.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6B6T
 
 | |
6B6O
 
 | |
6B6S
 
 | |
5YPC
 
 | | p62/SQSTM1 ZZ domain with Phe-peptide | | Descriptor: | 78 kDa glucose-regulated protein,Sequestosome-1, ZINC ION | | Authors: | Kwon, D.H, Kim, L, Song, H.K. | | Deposit date: | 2017-11-01 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.962 Å) | | Cite: | Insights into degradation mechanism of N-end rule substrates by p62/SQSTM1 autophagy adapter.
Nat Commun, 9, 2018
|
|
5YSK
 
 | | SdeA mART-C domain EE/AA apo | | Descriptor: | Ubiquitinating/deubiquitinating enzyme SdeA | | Authors: | Kim, L, Kwon, D.H, Song, H.K. | | Deposit date: | 2017-11-14 | | Release date: | 2018-08-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | Structural and Biochemical Study of the Mono-ADP-Ribosyltransferase Domain of SdeA, a Ubiquitylating/Deubiquitylating Enzyme from Legionella pneumophila
J. Mol. Biol., 430, 2018
|
|
2YIB
 
 | | Structure of the RNA polymerase VP1 from Infectious Pancreatic Necrosis Virus | | Descriptor: | RNA-DIRECTED RNA POLYMERASE | | Authors: | Graham, S.C, Sarin, L.P, Bahar, M.W, Myers, R.A, Stuart, D.I, Bamford, D.H, Grimes, J.M. | | Deposit date: | 2011-05-11 | | Release date: | 2011-07-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | The N-Terminus of the RNA Polymerase from Infectious Pancreatic Necrosis Virus is the Determinant of Genome Attachment.
Plos Pathog., 7, 2011
|
|
