4MAT
 
 | | E.COLI METHIONINE AMINOPEPTIDASE HIS79ALA MUTANT | | Descriptor: | PROTEIN (METHIONINE AMINOPEPTIDASE), SODIUM ION | | Authors: | Lowther, W.T, Orville, A.M, Madden, D.T, Lim, S, Rich, D.H, Matthews, B.W. | | Deposit date: | 1999-03-29 | | Release date: | 1999-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Escherichia coli methionine aminopeptidase: implications of crystallographic analyses of the native, mutant, and inhibited enzymes for the mechanism of catalysis.
Biochemistry, 38, 1999
|
|
1MV2
 
 | | The tandem, Face-to-Face AP Pairs in 5'(rGGCAPGCCU)2 | | Descriptor: | 5'-R(*GP*GP*CP*AP*(P5P)P*GP*CP*CP*U)-3' | | Authors: | Znosko, B.M, Burkard, M.E, Krugh, T.R, Turner, D.H. | | Deposit date: | 2002-09-24 | | Release date: | 2002-12-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Molecular Recognition in Purine-Rich Internal Loops: Thermodynamic, Structural, and Dynamic Consequences of Purine for Adenine Substitutions in 5'(rGGCAAGCCU)2
Biochemistry, 41, 2002
|
|
1MV6
 
 | | The tandem, Sheared PP Pairs in 5'(rGGCPPGCCU)2 | | Descriptor: | 5'-R(*GP*GP*CP*(P5P)P*(P5P)P*GP*CP*CP*U)-3' | | Authors: | Znosko, B.M, Burkard, M.E, Krugh, T.R, Turner, D.H. | | Deposit date: | 2002-09-24 | | Release date: | 2002-12-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Molecular Recognition in Purine-Rich Internal Loops: Thermodynamic, Structural, and Dynamic Consequences of Purine for Adenine Substitutions in 5'(rGGCAAGCCU)2
Biochemistry, 41, 2002
|
|
1DLQ
 
 | | STRUCTURE OF CATECHOL 1,2-DIOXYGENASE FROM ACINETOBACTER SP. ADP1 INHIBITED BY BOUND MERCURY | | Descriptor: | CATECHOL 1,2-DIOXYGENASE, FE (III) ION, MERCURY (II) ION, ... | | Authors: | Vetting, M.W, Ohlendorf, D.H. | | Deposit date: | 1999-12-11 | | Release date: | 2000-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The 1.8 A crystal structure of catechol 1,2-dioxygenase reveals a novel hydrophobic helical zipper as a subunit linker.
Structure Fold.Des., 8, 2000
|
|
1DP0
 
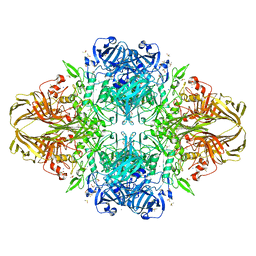 | | E. COLI BETA-GALACTOSIDASE AT 1.7 ANGSTROM | | Descriptor: | BETA-GALACTOSIDASE, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Juers, D.H, Jacobson, R.H, Wigley, D, Zhang, X.J, Huber, R.E, Tronrud, D.E, Matthews, B.W. | | Deposit date: | 1999-12-22 | | Release date: | 2001-02-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High resolution refinement of beta-galactosidase in a new crystal form reveals multiple metal-binding sites and provides a structural basis for alpha-complementation.
Protein Sci., 9, 2000
|
|
1HB9
 
 | | quasi-atomic resolution model of bacteriophage PRD1 wild type virion, obtained by combined cryo-EM and X-ray crystallography. | | Descriptor: | BACTERIOPHAGE PRD1 | | Authors: | San Martin, C, Burnett, R.M, De Haas, F, Heinkel, R, Rutten, T, Fuller, S.D, Butcher, S.J, Bamford, D.H. | | Deposit date: | 2001-04-13 | | Release date: | 2001-12-05 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (25 Å) | | Cite: | Combined Em/X-Ray Imaging Yields a Quasi-Atomic Model of the Adenovirus-Related Bacteriophage Prd1 and Shows Key Capsid and Membrane Interactions
Structure, 9, 2001
|
|
1DLT
 
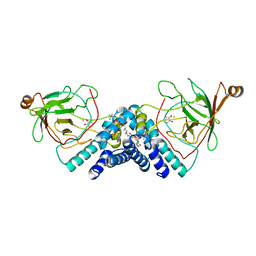 | | STRUCTURE OF CATECHOL 1,2-DIOXYGENASE FROM ACINETOBACTER SP. ADP1 WITH BOUND CATECHOL | | Descriptor: | CATECHOL, CATECHOL 1,2-DIOXYGENASE, FE (III) ION, ... | | Authors: | Vetting, M.W, Ohlendorf, D.H. | | Deposit date: | 1999-12-12 | | Release date: | 2000-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The 1.8 A crystal structure of catechol 1,2-dioxygenase reveals a novel hydrophobic helical zipper as a subunit linker.
Structure Fold.Des., 8, 2000
|
|
1HB5
 
 | | quasi-atomic resolution model of bacteriophage PRD1 P3-shell, obtained by combined cryo-EM and X-ray crystallography. | | Descriptor: | BACTERIOPHAGE PRD1 P3-SHELL | | Authors: | San Martin, C, Burnett, R.M, De Haas, F, Heinkel, R, Rutten, T, Fuller, S.D, Butcher, S.J, Bamford, D.H. | | Deposit date: | 2001-04-11 | | Release date: | 2001-12-05 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Combined Em/X-Ray Imaging Yields a Quasi-Atomic Model of the Adenovirus-Related Bacteriophage Prd1 and Shows Key Capsid and Membrane Interactions.
Structure, 9, 2001
|
|
1KYJ
 
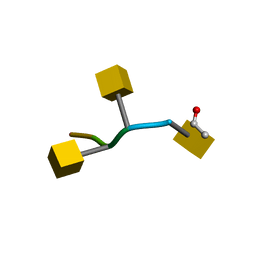 | | Tumor Associated Mucin Motif from CD43 protein | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, Leukosialin (CD43) fragment | | Authors: | Coltart, D.M, Williams, L.J, Glunz, P.W, Sames, D, Kuduk, S.D, Schwarz, J.B, Chen, X.-T, Royyuru, A.K, Danishefsky, S.D, Live, D.H. | | Deposit date: | 2002-02-04 | | Release date: | 2002-02-20 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | Principles of Mucin Architecture: Structural Studies on Synthetic Glycopeptides Bearing Clustered Mono-, Di-, Tri-, and Hexasaccharide Glycodomains
J.Am.Chem.Soc., 124, 2002
|
|
1HB7
 
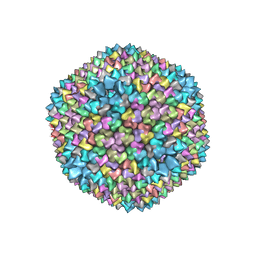 | | quasi-atomic resolution model of bacteriophage PRD1 sus1 mutant, obtained by combined cryo-EM and X-ray crystallography. | | Descriptor: | BACTERIOPHAGE PRD1 SUS1 MUTANT CAPSID | | Authors: | San Martin, C, Burnett, R.M, De Haas, F, Heinkel, R, Rutten, T, Fuller, S.D, Butcher, S.J, Bamford, D.H. | | Deposit date: | 2001-04-12 | | Release date: | 2001-12-05 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (14 Å) | | Cite: | Combined Em/X-Ray Imaging Yields a Quasi-Atomic Model of the Adenovirus-Related Bacteriophage Prd1 and Shows Key Capsid and Membrane Interactions.
Structure, 9, 2001
|
|
1ECY
 
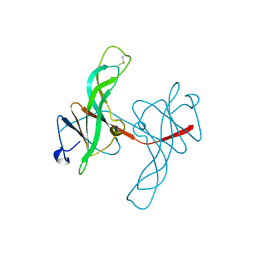 | | PROTEASE INHIBITOR ECOTIN | | Descriptor: | ECOTIN, alpha-D-glucopyranose, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose, ... | | Authors: | Shin, D.H, Suh, S.W. | | Deposit date: | 1996-08-06 | | Release date: | 1997-02-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal structure analyses of uncomplexed ecotin in two crystal forms: implications for its function and stability.
Protein Sci., 5, 1996
|
|
1NTT
 
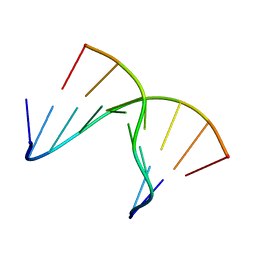 | | 5'(dCPCPUPCPCPUPUP)3':(rAGGAGGAAA)5', where P=propynyl | | Descriptor: | 5'-D(*CP*(5PC)P*(PDU)P*(5PC)P*(5PC)P*(PDU)P*(PDU))-3', 5'-R(*AP*AP*AP*GP*GP*AP*GP*GP*A)-3' | | Authors: | Znosko, B.M, Barnes III, T.W, Krugh, T.R, Turner, D.H. | | Deposit date: | 2003-01-30 | | Release date: | 2003-06-10 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR Studies of DNA Single Strands and DNA:RNA Hybrids With and Without 1-Propynylation at C5 of Oligopyrimidines
J.Am.Chem.Soc., 125, 2003
|
|
1CJD
 
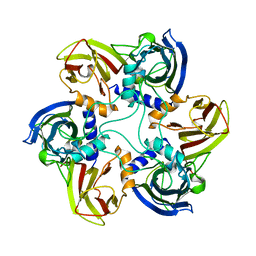 | | THE BACTERIOPHAGE PRD1 COAT PROTEIN, P3, IS STRUCTURALLY SIMILAR TO HUMAN ADENOVIRUS HEXON | | Descriptor: | PROTEIN (MAJOR CAPSID PROTEIN (P3)) | | Authors: | Benson, S.D, Bamford, J.K.H, Bamford, D.H, Burnett, R.M. | | Deposit date: | 1999-04-12 | | Release date: | 1999-09-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Viral evolution revealed by bacteriophage PRD1 and human adenovirus coat protein structures.
Cell(Cambridge,Mass.), 98, 1999
|
|
1ECZ
 
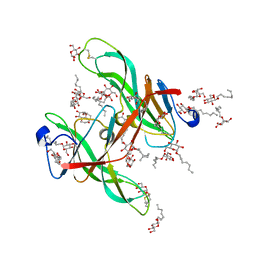 | | PROTEASE INHIBITOR ECOTIN | | Descriptor: | ECOTIN, octyl beta-D-glucopyranoside | | Authors: | Shin, D.H, Suh, S.W. | | Deposit date: | 1996-08-06 | | Release date: | 1997-02-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Crystal structure analyses of uncomplexed ecotin in two crystal forms: implications for its function and stability.
Protein Sci., 5, 1996
|
|
1NTS
 
 | | 5'(dCCPUPCPCPUPUP)3':3'(rAGGAGGAAA)5', where P=propynyl | | Descriptor: | 5'-D(*(5PC)P*(5PC)P*(PDU)P*(5PC)P*(5PC)P*(PDU)P*(PDU))-3', 5'-R(*AP*AP*AP*GP*GP*AP*GP*GP*A)-3' | | Authors: | Znosko, B.M, Barnes III, T.W, Krugh, T.R, Turner, D.H. | | Deposit date: | 2003-01-30 | | Release date: | 2003-06-10 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR Studies of DNA Single Strands and DNA:RNA Hybrids With and Without 1-Propynylation at C5 of Oligopyrimidines
J.Am.Chem.Soc., 125, 2003
|
|
1ODO
 
 | | 1.85 A structure of CYP154A1 from Streptomyces coelicolor A3(2) | | Descriptor: | 4-PHENYL-1H-IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE, PUTATIVE CYTOCHROME P450 154A1 | | Authors: | Podust, L.M, Kim, Y, Arase, M, Bach, H, Sherman, D.H, Lamb, D.C, Kelly, S.L, Waterman, M.R. | | Deposit date: | 2003-02-19 | | Release date: | 2004-01-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Comparison of the 1.85 A Structure of Cyp154A1 from Streptomyces Coelicolor A3(2) with the Closely Related Cyp154C1 and Cyps from Antibiotic Biosynthetic Pathways.
Protein Sci., 13, 2004
|
|
1NTQ
 
 | | 5'(dCCUCCUU)3':3'(rAGGAGGAAA)5' | | Descriptor: | 5'-D(*CP*CP*UP*CP*CP*UP*U)-3', 5'-R(*AP*AP*AP*GP*GP*AP*GP*GP*A)-3' | | Authors: | Znosko, B.M, Barnes III, T.W, Krugh, T.R, Turner, D.H. | | Deposit date: | 2003-01-30 | | Release date: | 2003-06-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Studies of DNA Single Strands and DNA:RNA Hybrids With and Without 1-Propynylation at C5 of Oligopyrimidines
J.Am.Chem.Soc., 125, 2003
|
|
1DMH
 
 | | STRUCTURE OF CATECHOL 1,2-DIOXYGENASE FROM ACINETOBACTER SP. ADP1 WITH BOUND 4-METHYLCATECHOL | | Descriptor: | 4-METHYLCATECHOL, CATECHOL 1,2-DIOXYGENASE, FE (III) ION, ... | | Authors: | Vetting, M.W, Ohlendorf, D.H. | | Deposit date: | 1999-12-14 | | Release date: | 2000-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The 1.8 A crystal structure of catechol 1,2-dioxygenase reveals a novel hydrophobic helical zipper as a subunit linker.
Structure Fold.Des., 8, 2000
|
|
1DLM
 
 | | STRUCTURE OF CATECHOL 1,2-DIOXYGENASE FROM ACINETOBACTER CALCOACETICUS NATIVE DATA | | Descriptor: | CATECHOL 1,2-DIOXYGENASE, FE (III) ION, [1-PENTADECANOYL-2-DECANOYL-GLYCEROL-3-YL]PHOSPHONYL CHOLINE | | Authors: | Vetting, M.W, Ohlendorf, D.H. | | Deposit date: | 1999-12-11 | | Release date: | 2000-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 1.8 A crystal structure of catechol 1,2-dioxygenase reveals a novel hydrophobic helical zipper as a subunit linker.
Structure Fold.Des., 8, 2000
|
|
1MUV
 
 | | Sheared A(anti)-A(anti) Base Pairs in a Destabilizing 2x2 Internal Loop: The NMR Structure of 5'(rGGCAAGCCU)2 | | Descriptor: | 5'-R(*GP*GP*CP*AP*AP*GP*CP*CP*U)-3' | | Authors: | Znosko, B.M, Burkard, M.E, Schroeder, S.J, Krugh, T.R, Turner, D.H. | | Deposit date: | 2002-09-24 | | Release date: | 2002-12-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Sheared Aanti-Aanti Base Pairs in a Destabilizing 2x2 Internal Loop: The NMR Structure of
5'(rGGCAAGCCU)2
Biochemistry, 41, 2002
|
|
1MV1
 
 | | The Tandem, Sheared PA Pairs in 5'(rGGCPAGCCU)2 | | Descriptor: | 5'-R(*GP*GP*CP*(P5P)P*AP*GP*CP*CP*U)-3' | | Authors: | Znosko, B.M, Burkard, M.E, Krugh, T.R, Turner, D.H. | | Deposit date: | 2002-09-24 | | Release date: | 2002-12-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Molecular Recognition in Purine-Rich Internal Loops: Thermodynamic, Structural, and Dynamic Consequences of Purine for Adenine Substitutions in 5'(rGGCAAGCCU)2
Biochemistry, 41, 2002
|
|
1TS5
 
 | | I140T MUTANT OF TOXIC SHOCK SYNDROME TOXIN-1 FROM S. AUREUS | | Descriptor: | TOXIC SHOCK SYNDROME TOXIN-1 | | Authors: | Earhart, C.A, Mitchell, D.T, Murray, D.L, Pinheiro, D.M, Matsumura, M, Schlievert, P.M, Ohlendorf, D.H. | | Deposit date: | 1997-10-10 | | Release date: | 1998-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of five mutants of toxic shock syndrome toxin-1 with reduced biological activity.
Biochemistry, 37, 1998
|
|
5NMR
 
 | | Monomeric mouse Sortilin extracellular domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Leloup, N.O.L, Loessl, P, Meijer, D.H.M, Heck, A.J.R, Thies-Weesie, D.M.E, Janssen, B.J.C. | | Deposit date: | 2017-04-07 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Low pH-induced conformational change and dimerization of sortilin triggers endocytosed ligand release.
Nat Commun, 8, 2017
|
|
5NOJ
 
 | | Ca2+-induced Movement of Tropomyosin on Native Cardiac Thin Filaments - "OPEN" state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Risi, C, Eisner, J, Belknap, B, Heeley, D.H, White, H.D, Schroeder, G.F, Galkin, V.E. | | Deposit date: | 2017-04-12 | | Release date: | 2017-08-02 | | Last modified: | 2017-08-09 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Ca(2+)-induced movement of tropomyosin on native cardiac thin filaments revealed by cryoelectron microscopy.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5NOL
 
 | | Ca2+-induced Movement of Tropomyosin on Native Cardiac Thin Filaments - "Closed" state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cardiac muscle alpha actin 1, MAGNESIUM ION, ... | | Authors: | Risi, C, Eisner, J, Belknap, B, Heeley, D.H, White, H.D, Schroeder, G.F, Galkin, V.E. | | Deposit date: | 2017-04-12 | | Release date: | 2017-07-19 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Ca(2+)-induced movement of tropomyosin on native cardiac thin filaments revealed by cryoelectron microscopy.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
