2MXL
 
 | |
2L8P
 
 | | Solution Structure of a DNA Duplex Containing the Potent Anti-Poxvirus Agent Cidofovir | | Descriptor: | DNA (5'-D(*CP*GP*CP*AP*TP*GP*(L8P)P*TP*AP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*TP*AP*GP*CP*AP*TP*GP*CP*G)-3') | | Authors: | Julien, O, Beadle, J.R, Magee, W.C, Chatterjee, S, Hostetler, K.Y, Evans, D.H, Sykes, B.D. | | Deposit date: | 2011-01-22 | | Release date: | 2011-02-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a DNA duplex containing the potent anti-poxvirus agent cidofovir.
J.Am.Chem.Soc., 133, 2011
|
|
1IK4
 
 | | X-ray Structure of Methylglyoxal Synthase from E. coli Complexed with Phosphoglycolohydroxamic Acid | | Descriptor: | METHYLGLYOXAL SYNTHASE, PHOSPHOGLYCOLOHYDROXAMIC ACID | | Authors: | Marks, G.T, Harris, T.K, Massiah, M.A, Mildvan, A.S, Harrison, D.H.T. | | Deposit date: | 2001-05-02 | | Release date: | 2001-09-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanistic implications of methylglyoxal synthase complexed with phosphoglycolohydroxamic acid as observed by X-ray crystallography and NMR spectroscopy.
Biochemistry, 40, 2001
|
|
2MH2
 
 | | Structural insights into the DNA recognition and protein interaction domains reveal fundamental homologous DNA pairing properties of HOP2 | | Descriptor: | Homologous-pairing protein 2 homolog | | Authors: | Moktan, H, Guiraldelli, M.F, Eyter, C.A, Zhao, W, Camerini-Otero, R.D, Sung, P, Zhou, D.H, Pezza, R.J. | | Deposit date: | 2013-11-13 | | Release date: | 2014-04-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and DNA-binding Properties of the Winged Helix Domain of the Meiotic Recombination HOP2 Protein.
J.Biol.Chem., 289, 2014
|
|
2K89
 
 | | Solution structure of a novel Ubiquitin-binding domain from Human PLAA (PFUC, Gly76-Pro77 cis isomer) | | Descriptor: | Phospholipase A-2-activating protein | | Authors: | Fu, Q.S, Zhou, C.J, Gao, H.C, Lin, D.H, Hu, H.Y. | | Deposit date: | 2008-09-04 | | Release date: | 2009-05-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Ubiquitin Recognition by a Novel Domain from Human Phospholipase A2-activating Protein.
J.Biol.Chem., 284, 2009
|
|
2K8B
 
 | | Solution structure of PLAA family ubiquitin binding domain (PFUC) cis isomer in complex with ubiquitin | | Descriptor: | Phospholipase A-2-activating protein, Ubiquitin | | Authors: | Fu, Q.S, Zhou, C.J, Gao, H.C, Lin, D.H, Hu, H.Y. | | Deposit date: | 2008-09-04 | | Release date: | 2009-05-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Ubiquitin Recognition by a Novel Domain from Human Phospholipase A2-activating Protein.
J.Biol.Chem., 284, 2009
|
|
2K8A
 
 | | Solution structure of a novel Ubiquitin-binding domain from Human PLAA (PFUC, Gly76-Pro77 trans isomer) | | Descriptor: | Phospholipase A-2-activating protein | | Authors: | Fu, Q.S, Zhou, C.J, Gao, H.C, Lin, D.H, Hu, H.Y. | | Deposit date: | 2008-09-04 | | Release date: | 2009-05-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Ubiquitin Recognition by a Novel Domain from Human Phospholipase A2-activating Protein.
J.Biol.Chem., 284, 2009
|
|
2O81
 
 | |
2O83
 
 | |
2NZ1
 
 | |
2KDY
 
 | | NMR structure of LP2086-B01 | | Descriptor: | Factor H binding protein variant B01_001 | | Authors: | Mascioni, A, Bentley, B.E, Camarda, R, Dilts, D.A, Fink, P, Gusarova, V, Hoiseth, S, Jacob, J, Lin, S.L, Malakian, K, McNeil, L.K, Mininni, T, Moy, F, Murphy, E, Novikova, E, Sigethy, S, Wen, Y, Zlotnick, G.W, Tsao, D.H.H. | | Deposit date: | 2009-01-20 | | Release date: | 2009-02-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for the Immunogenic Properties of the Meningococcal Vaccine Candidate LP2086.
J.Biol.Chem., 284, 2009
|
|
2MIZ
 
 | |
2O5N
 
 | | Crystal structure of a Viral Glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MuHV1gpm153, ... | | Authors: | Mans, J, Natarajan, K, Robinson, H, Margulies, D.H. | | Deposit date: | 2006-12-06 | | Release date: | 2007-09-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Cellular Expression and Crystal Structure of the Murine Cytomegalovirus Major Histocompatibility Complex Class I-like Glycoprotein, m153.
J.Biol.Chem., 282, 2007
|
|
2MAT
 
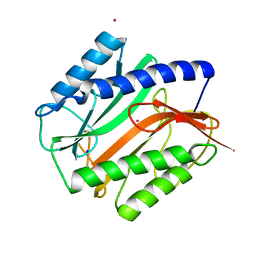 | | E.COLI METHIONINE AMINOPEPTIDASE AT 1.9 ANGSTROM RESOLUTION | | Descriptor: | COBALT (II) ION, PROTEIN (METHIONINE AMINOPEPTIDASE), SODIUM ION | | Authors: | Lowther, W.T, Orville, A.M, Madden, D.T, Lim, S, Rich, D.H, Matthews, B.W. | | Deposit date: | 1999-03-29 | | Release date: | 1999-06-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Escherichia coli methionine aminopeptidase: implications of crystallographic analyses of the native, mutant, and inhibited enzymes for the mechanism of catalysis.
Biochemistry, 38, 1999
|
|
2LX1
 
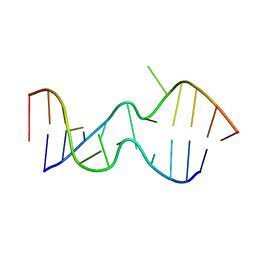 | |
2NYZ
 
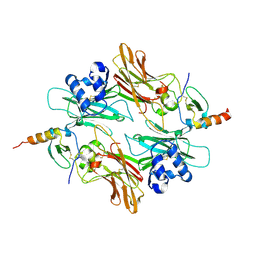 | |
2L8F
 
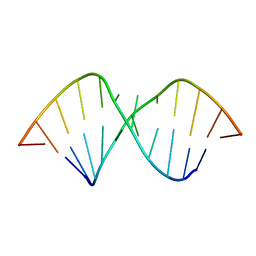 | |
2LIU
 
 | | NMR structure of holo-ACPI domain from CurA module from Lyngbya majuscula | | Descriptor: | CurA | | Authors: | Busche, A.E, Gottstein, D, Hein, C, Ripin, N, Pader, I, Tufar, P, Eisman, E.B, Gu, L, Walsh, C.T, Loehr, F, Sherman, D.H, Guntert, P, Dotsch, V. | | Deposit date: | 2011-09-01 | | Release date: | 2011-12-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Characterization of Molecular Interactions between ACP and Halogenase Domains in the Curacin A Polyketide Synthase.
Acs Chem.Biol., 7, 2012
|
|
1QWX
 
 | | Crystal Structure of a Staphylococcal Inhibitor/Chaperone | | Descriptor: | cysteine protease | | Authors: | Brown, C.K, Gu, Z.-Y, Nickerson, N, McGavin, M.J, Ohlendorf, D.H, Earhart, C.A. | | Deposit date: | 2003-09-03 | | Release date: | 2004-02-10 | | Last modified: | 2014-03-12 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: |
|
|
1JXE
 
 | |
1PIH
 
 | | THE THREE DIMENSIONAL STRUCTURE OF THE PARAMAGNETIC PROTEIN HIPIP I FROM E.HALOPHILA THROUGH NUCLEAR MAGNETIC RESONANCE | | Descriptor: | HIGH POTENTIAL IRON SULFUR PROTEIN, IRON/SULFUR CLUSTER | | Authors: | Banci, L, Bertini, I, Eltis, L.D, Felli, I, Kastrau, D.H.W, Luchinat, C, Piccioli, M, Pierattelli, R, Smith, M. | | Deposit date: | 1994-08-03 | | Release date: | 1994-12-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional structure in solution of the paramagnetic high-potential iron-sulfur protein I from Ectothiorhodospira halophila through nuclear magnetic resonance.
Eur.J.Biochem., 225, 1994
|
|
1PIJ
 
 | | THE THREE DIMENSIONAL STRUCTURE OF THE PARAMAGNETIC PROTEIN HIPIP I FROM E.HALOPHILA THROUGH NUCLEAR MAGNETIC RESONANCE | | Descriptor: | HIGH POTENTIAL IRON SULFUR PROTEIN, IRON/SULFUR CLUSTER | | Authors: | Banci, L, Bertini, I, Eltis, L.D, Felli, I.C, Kastrau, D.H.W, Luchinat, C, Piccioli, M, Pierattelli, R, Smith, M. | | Deposit date: | 1994-11-11 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional structure in solution of the paramagnetic high-potential iron-sulfur protein I from Ectothiorhodospira halophila through nuclear magnetic resonance.
Eur.J.Biochem., 225, 1994
|
|
1S8G
 
 | | Crystal structure of Lys49-Phospholipase A2 from Agkistrodon contortrix laticinctus, fatty acid bound form | | Descriptor: | GLYCEROL, LAURIC ACID, Phospholipase A2 homolog, ... | | Authors: | Ambrosio, A.L.B, de Souza, D.H.F, Nonato, M.C, Selistre de Araujo, H.S, Ownby, C.L, Garratt, R.C. | | Deposit date: | 2004-02-02 | | Release date: | 2004-02-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Molecular Mechanism for Lys49-Phospholipase A2 Activity Based on Ligand-induced Conformational Change.
J.Biol.Chem., 280, 2005
|
|
1S8I
 
 | | Crystal structure of Lys49-Phospholipase A2 from Agkistrodon contortrix laticinctus, second fatty acid free form | | Descriptor: | Phospholipase A2 homolog, SULFATE ION | | Authors: | Ambrosio, A.L.B, de Souza, D.H.F, Nonato, M.C, Selistre de Araujo, H.S, Ownby, C.L, Garratt, R.C. | | Deposit date: | 2004-02-02 | | Release date: | 2004-02-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.609 Å) | | Cite: | A Molecular Mechanism for Lys49-Phospholipase A2 Activity Based on Ligand-induced Conformational Change.
J.Biol.Chem., 280, 2005
|
|
1S8H
 
 | | Crystal structure of Lys49-Phospholipase A2 from Agkistrodon contortrix laticinctus, first fatty acid free form | | Descriptor: | Phospholipase A2 homolog, SULFATE ION | | Authors: | Ambrosio, A.L.B, de Souza, D.H.F, Nonato, M.C, Selistre de Araujo, H.S, Ownby, C.L, Garratt, R.C. | | Deposit date: | 2004-02-02 | | Release date: | 2004-02-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Molecular Mechanism for Lys49-Phospholipase A2 Activity Based on Ligand-induced Conformational Change.
J.Biol.Chem., 280, 2005
|
|
