4QDX
 
 | | Crystal structure of Antigen 85C-C209G mutant | | Descriptor: | Diacylglycerol acyltransferase/mycolyltransferase Ag85C, SULFATE ION | | Authors: | Favrot, L, Lajiness, D.H, Ronning, D.R. | | Deposit date: | 2014-05-14 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.503 Å) | | Cite: | Inactivation of the Mycobacterium tuberculosis Antigen 85 Complex by Covalent, Allosteric Inhibitors.
J.Biol.Chem., 289, 2014
|
|
4QEK
 
 | | Crystal structure of Antigen 85C-S124A mutant | | Descriptor: | Diacylglycerol acyltransferase/mycolyltransferase Ag85C, SULFATE ION, alpha-D-glucopyranose | | Authors: | Favrot, L, Lajiness, D.H, Ronning, D.R. | | Deposit date: | 2014-05-16 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.299 Å) | | Cite: | Inactivation of the Mycobacterium tuberculosis Antigen 85 Complex by Covalent, Allosteric Inhibitors.
J.Biol.Chem., 289, 2014
|
|
4QDZ
 
 | | Crystal structure of Antigen 85C-E228Q mutant | | Descriptor: | ACETATE ION, Diacylglycerol acyltransferase/mycolyltransferase Ag85C, HEXAETHYLENE GLYCOL | | Authors: | Favrot, L, Lajiness, D.H, Ronning, D.R. | | Deposit date: | 2014-05-14 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.885 Å) | | Cite: | Inactivation of the Mycobacterium tuberculosis Antigen 85 Complex by Covalent, Allosteric Inhibitors.
J.Biol.Chem., 289, 2014
|
|
4QBU
 
 | | Structure of the Acyl Transferase domain of ZmaA | | Descriptor: | FORMIC ACID, ZmaA | | Authors: | Dyer, D.H, Kevany, B.M, Thomas, M.G, Forest, K.T. | | Deposit date: | 2014-05-08 | | Release date: | 2014-11-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Polyketide Synthase Acyltransferase Domain Structure Suggests a Recognition Mechanism for Its Hydroxymalonyl-Acyl Carrier Protein Substrate.
Plos One, 9, 2014
|
|
4P87
 
 | | Crystal structure of Est-Y29, a novel penicillin-binding protein/beta-lactamase homolog from a metagenomic library | | Descriptor: | 4-NITROPHENYL PHOSPHATE, Est-Y29 | | Authors: | Ngo, T.D, Ryu, B.H, Ju, H.S, Jang, E.J, Kim, K.K, Kim, D.H. | | Deposit date: | 2014-03-30 | | Release date: | 2014-09-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Crystallographic analysis and biochemical applications of a novel penicillin-binding protein/ beta-lactamase homologue from a metagenomic library.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4QDO
 
 | |
4QDU
 
 | |
4QE3
 
 | | Crystal structure of Antigen 85C-H260Q mutant | | Descriptor: | ACETIC ACID, CHAPSO, Diacylglycerol acyltransferase/mycolyltransferase Ag85C | | Authors: | Favrot, L, Lajiness, D.H, Ronning, D.R. | | Deposit date: | 2014-05-15 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.351 Å) | | Cite: | Inactivation of the Mycobacterium tuberculosis Antigen 85 Complex by Covalent, Allosteric Inhibitors.
J.Biol.Chem., 289, 2014
|
|
5FOI
 
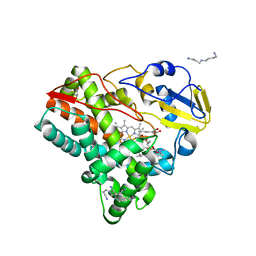 | | Crystal structure of mycinamicin VIII C21 methyl hydroxylase MycCI from Micromonospora griseorubida bound to mycinamicin VIII | | Descriptor: | GLYCEROL, MYCINAMICIN VIII C21 METHYL HYDROXYLASE, Mycinamicin VIII, ... | | Authors: | Demars, M, Sheng, F, Podust, L.M, Sherman, D.H. | | Deposit date: | 2015-11-22 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Biochemical and Structural Characterization of Mycci, a Versatile P450 Biocatalyst from the Mycinamicin Biosynthetic Pathway.
Acs Chem.Biol., 11, 2016
|
|
5GRZ
 
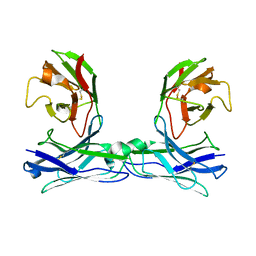 | | Crystal structure of disulfide-bonded diabody | | Descriptor: | diabody | | Authors: | Kim, J.H, Song, D.H, Youn, S.J, Kim, J.W, Cho, G, Lee, H, Lee, J.O. | | Deposit date: | 2016-08-13 | | Release date: | 2016-10-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of mono- and bi-specific diabodies and reduction of their structural flexibility by introduction of disulfide bridges at the Fv interface.
Sci Rep, 6, 2016
|
|
5GAI
 
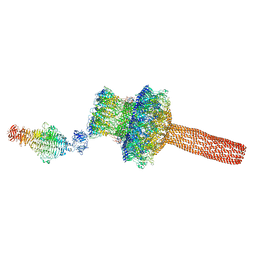 | | Probabilistic Structural Models of Mature P22 Bacteriophage Portal, Hub, and Tailspike proteins | | Descriptor: | Peptidoglycan hydrolase gp4, Portal protein, Tail fiber protein | | Authors: | Pintilie, G, Chen, D.H, Haase-Pettingell, C.A, King, J.A, Chiu, W. | | Deposit date: | 2015-12-01 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (10.5 Å) | | Cite: | Resolution and Probabilistic Models of Components in CryoEM Maps of Mature P22 Bacteriophage.
Biophys.J., 110, 2016
|
|
5GS2
 
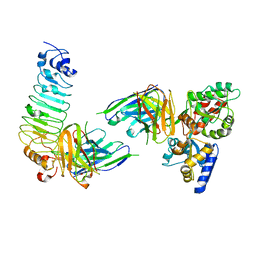 | | Crystal structure of diabody complex with repebody and MBP | | Descriptor: | Maltose-binding periplasmic protein, anti-MBP, anti-repebody, ... | | Authors: | Kim, J.H, Song, D.H, Youn, S.J, Kim, J.W, Cho, G, Lee, H, Lee, J.O. | | Deposit date: | 2016-08-13 | | Release date: | 2016-10-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.592 Å) | | Cite: | Crystal structure of mono- and bi-specific diabodies and reduction of their structural flexibility by introduction of disulfide bridges at the Fv interface.
Sci Rep, 6, 2016
|
|
5GRY
 
 | | Crystal structure of disulfide-bonded diabody | | Descriptor: | diabody | | Authors: | Kim, J.H, Song, D.H, Youn, S.J, Kim, J.W, Cho, G, Lee, H, Lee, J.O. | | Deposit date: | 2016-08-12 | | Release date: | 2016-10-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.639 Å) | | Cite: | Crystal structure of mono- and bi-specific diabodies and reduction of their structural flexibility by introduction of disulfide bridges at the Fv interface.
Sci Rep, 6, 2016
|
|
2BUR
 
 | | Crystal Structure Of Wild-Type Protocatechuate 3,4-Dioxygenase from Acinetobacter Sp. ADP1 in Complex with 4-hydroxybenzoate | | Descriptor: | FE (III) ION, P-HYDROXYBENZOIC ACID, PROTOCATECHUATE 3,4-DIOXYGENASE ALPHA CHAIN, ... | | Authors: | Vetting, M.W, Valley, M.P, D'Argenio, D.A, Ornston, L.N, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 2005-06-17 | | Release date: | 2006-09-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biophysical Analyses of Designed and Selected Mutants of Protocatechuate 3,4-Dioxygenase
Annu.Rev.Microbiol., 58, 2004
|
|
5GRU
 
 | | Structure of mono-specific diabody | | Descriptor: | Maltose-binding periplasmic protein, diabody protein | | Authors: | Kim, J.H, Song, D.H, Youn, S.J, Cho, G, Lee, H, Lee, J.O. | | Deposit date: | 2016-08-12 | | Release date: | 2016-10-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.955 Å) | | Cite: | Crystal structure of mono- and bi-specific diabodies and reduction of their structural flexibility by introduction of disulfide bridges at the Fv interface.
Sci Rep, 6, 2016
|
|
5GS1
 
 | | Crystal structure of homo-specific diabody | | Descriptor: | diabody, heavy chain, light chain | | Authors: | Kim, J.H, Song, D.H, Youn, S.J, Kim, J.W, Cho, G, Lee, H, Lee, J.O. | | Deposit date: | 2016-08-13 | | Release date: | 2016-10-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of mono- and bi-specific diabodies and reduction of their structural flexibility by introduction of disulfide bridges at the Fv interface.
Sci Rep, 6, 2016
|
|
5GRW
 
 | | Crystal structure of homo-specific diabody | | Descriptor: | homo-specific diabody protein | | Authors: | Kim, J.H, Song, D.H, Youn, S.J, Kim, J.W, Cho, G, Lee, H, Lee, J.O. | | Deposit date: | 2016-08-12 | | Release date: | 2016-10-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of mono- and bi-specific diabodies and reduction of their structural flexibility by introduction of disulfide bridges at the Fv interface.
Sci Rep, 6, 2016
|
|
5GS0
 
 | | Crystal structure of the complex of TLR3 and bi-specific diabody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Toll-like receptor 3, alpha-D-mannopyranose, ... | | Authors: | Kim, J.H, Song, D.H, Youn, S.J, Kim, J.W, Cho, G, Lee, H, Lee, J.O. | | Deposit date: | 2016-08-13 | | Release date: | 2016-10-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.275 Å) | | Cite: | Crystal structure of mono- and bi-specific diabodies and reduction of their structural flexibility by introduction of disulfide bridges at the Fv interface.
Sci Rep, 6, 2016
|
|
5GRV
 
 | | Crystal structure of homo-specific diabody | | Descriptor: | homo-specific diabody heavy chain, homo-specific diabody light chain | | Authors: | Kim, J.H, Song, D.H, Youn, S.J, Kim, J.W, Cho, G, Lee, H, Lee, J.O. | | Deposit date: | 2016-08-12 | | Release date: | 2016-10-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of mono- and bi-specific diabodies and reduction of their structural flexibility by introduction of disulfide bridges at the Fv interface.
Sci Rep, 6, 2016
|
|
2BUM
 
 | | Crystal Structure Of Wild-Type Protocatechuate 3,4-Dioxygenase from Acinetobacter Sp. ADP1 | | Descriptor: | FE (III) ION, HYDROXIDE ION, PROTOCATECHUATE 3,4-DIOXYGENASE ALPHA CHAIN, ... | | Authors: | Vetting, M.W, Valley, M.P, D'Argenio, D.A, Ornston, L.N, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 2005-06-14 | | Release date: | 2006-09-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biophysical Analyses of Designed and Selected Mutants of Protocatechuate 3,4-Dioxygenase
Annu.Rev.Microbiol., 58, 2004
|
|
5GS3
 
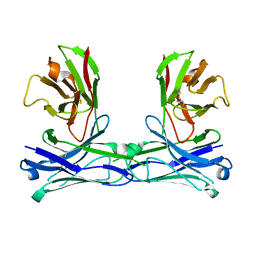 | | Crystal structure of diabody | | Descriptor: | diabody protein | | Authors: | Kim, J.H, Song, D.H, Youn, S.J, Kim, J.W, Cho, G, Lee, H, Lee, J.O. | | Deposit date: | 2016-08-13 | | Release date: | 2016-10-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Crystal structure of mono- and bi-specific diabodies and reduction of their structural flexibility by introduction of disulfide bridges at the Fv interface.
Sci Rep, 6, 2016
|
|
5GRX
 
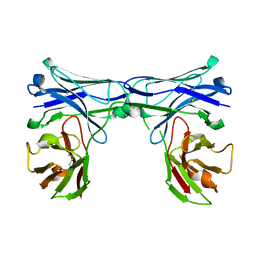 | | Crystal structure of disulfide-bonded diabody | | Descriptor: | diabody protein | | Authors: | Kim, J.H, Song, D.H, Youn, S.J, Kim, J.W, Cho, G, Lee, H, Lee, J.O. | | Deposit date: | 2016-08-12 | | Release date: | 2016-10-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Crystal structure of mono- and bi-specific diabodies and reduction of their structural flexibility by introduction of disulfide bridges at the Fv interface.
Sci Rep, 6, 2016
|
|
2BUW
 
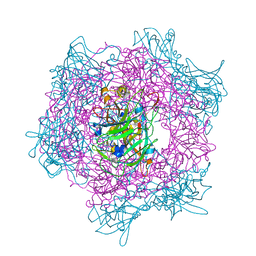 | | Crystal Structure of Protocatechuate 3,4-Dioxygenase from Acinetobacter Sp. ADP1 Mutant R457S in Complex with 4-Hydroxybenzoate | | Descriptor: | FE (III) ION, P-HYDROXYBENZOIC ACID, PROTOCATECHUATE 3,4-DIOXYGENASE ALPHA CHAIN, ... | | Authors: | Vetting, M.W, Valley, M.P, D'Argenio, D.A, Ornston, L.N, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 2005-06-20 | | Release date: | 2006-10-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic Studies of Intradiol Dioxygenases.
Phd Thesis, 2001
|
|
2C6H
 
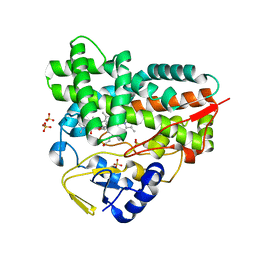 | | Crystal structure of YC-17-bound cytochrome P450 PikC (CYP107L1) | | Descriptor: | 4-{[4-(DIMETHYLAMINO)-3-HYDROXY-6-METHYLTETRAHYDRO-2H-PYRAN-2-YL]OXY}-12-ETHYL-3,5,7,11-TETRAMETHYLOXACYCLODODEC-9-ENE-2,8-DIONE, CYTOCHROME P450 MONOOXYGENASE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Sherman, D.H, Li, S, Yermalitskaya, L.V, Kim, Y, Smith, J.A, Waterman, M.R, Podust, L.M. | | Deposit date: | 2005-11-09 | | Release date: | 2006-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Structural Basis for Substrate Anchoring, Active Site Selectivity, and Product Formation by P450 Pikc from Streptomyces Venezuelae.
J.Biol.Chem., 281, 2006
|
|
2BUQ
 
 | | Crystal Structure Of Wild-Type Protocatechuate 3,4-Dioxygenase from Acinetobacter Sp. ADP1 in Complex with Catechol | | Descriptor: | CATECHOL, FE (III) ION, PROTOCATECHUATE 3,4-DIOXYGENASE ALPHA CHAIN, ... | | Authors: | Vetting, M.W, Valley, M.P, D'Argenio, D.A, Ornston, L.N, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 2005-06-17 | | Release date: | 2006-09-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biophysical Analyses of Designed and Selected Mutants of Protocatechuate 3,4-Dioxygenase
Annu.Rev.Microbiol., 58, 2004
|
|
