5WPP
 
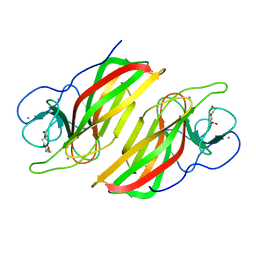 | | Crystal structure HpiC1 W73M/K132M | | Descriptor: | 12-epi-hapalindole C/U synthase, CALCIUM ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Newmister, S.A, Li, S, Garcia-Borras, M, Sanders, J.N, Yang, S, Lowell, A.N, Yu, F, Smith, J.L, Williams, R.M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2017-08-07 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of the Cope rearrangement and cyclization in hapalindole biogenesis.
Nat. Chem. Biol., 14, 2018
|
|
5WIS
 
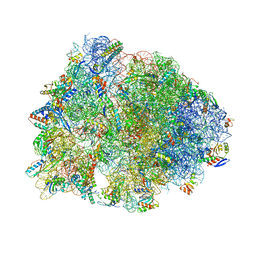 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with methymycin and bound to mRNA and A-, P- and E-site tRNAs at 2.7A resolution | | Descriptor: | (3R,4S,5S,7R,9E,11S,12R)-12-ethyl-11-hydroxy-3,5,7,11-tetramethyl-2,8-dioxooxacyclododec-9-en-4-yl 3,4,6-trideoxy-3-(dimethylamino)-beta-D-xylo-hexopyranoside, 16S Ribosomal RNA, 23S ribosomal RNA, ... | | Authors: | Almutairi, M.M, Svetlov, M.S, Hansen, D.A, Khabibullina, N.F, Klepacki, D, Kang, H.Y, Sherman, D.H, Vazquez-Laslop, N, Polikanov, Y.S, Mankin, A.S. | | Deposit date: | 2017-07-20 | | Release date: | 2018-02-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Co-produced natural ketolides methymycin and pikromycin inhibit bacterial growth by preventing synthesis of a limited number of proteins.
Nucleic Acids Res., 45, 2017
|
|
4P6B
 
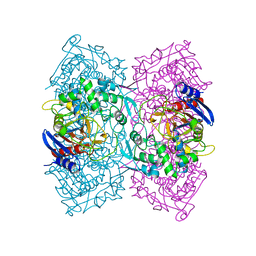 | | Crystal structure of Est-Y29,a novel penicillin-binding protein/beta-lactamase homolog from a metagenomic library | | Descriptor: | Est-Y29 | | Authors: | Ngo, T.D, Ryu, B.H, Ju, H.S, Jang, E.J, Kim, K.K, Kim, D.H. | | Deposit date: | 2014-03-24 | | Release date: | 2014-09-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystallographic analysis and biochemical applications of a novel penicillin-binding protein/ beta-lactamase homologue from a metagenomic library.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4PNH
 
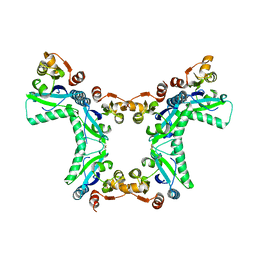 | |
4PR3
 
 | | Crystal structure of Brucella melitensis 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase | | Descriptor: | 5'-methylthioadenosine nucleosidase / s-adenosylhomocysteine nucleosidase, ADENINE, GLYCEROL, ... | | Authors: | Zhang, X.C, Kang, X.S, Zhao, Y, Jiang, D.H, Li, X.M, Chen, Z.L. | | Deposit date: | 2014-03-05 | | Release date: | 2014-04-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.606 Å) | | Cite: | Crystal structure and biochemical studies of Brucella melitensis 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase
Biochem.Biophys.Res.Commun., 446, 2014
|
|
3IXY
 
 | | The pseudo-atomic structure of dengue immature virus in complex with Fab fragments of the anti-fusion loop antibody E53 | | Descriptor: | E53 Fab Fragment (chain H), E53 Fab Fragment (chain L), Envelope protein E, ... | | Authors: | Cherrier, M.V, Kaufmann, B, Nybakken, G.E, Lok, S.M, Warren, J.T, Nelson, C.A, Kostyuchenko, V.A, Holdaway, H.A, Chipman, P.R, Kuhn, R.J, Diamond, M.S, Rossmann, M.G, Fremont, D.H. | | Deposit date: | 2009-02-26 | | Release date: | 2009-10-27 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (23 Å) | | Cite: | Structural basis for the preferential recognition of immature flaviviruses by a fusion-loop antibody
Embo J., 28, 2009
|
|
5UUB
 
 | |
5WIT
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with pikromycin and bound to mRNA and A-, P- and E-site tRNAs at 2.6A resolution | | Descriptor: | (3R,5R,6S,7S,9R,11E,13S,14R)-14-ethyl-13-hydroxy-3,5,7,9,13-pentamethyl-2,4,10-trioxo-1-oxacyclotetradec-11-en-6-yl 3,4,6-trideoxy-3-(dimethylamino)-beta-D-xylo-hexopyranoside, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Almutairi, M.M, Svetlov, M.S, Hansen, D.A, Khabibullina, N.F, Klepacki, D, Kang, H.Y, Sherman, D.H, Vazquez-Laslop, N, Polikanov, Y.S, Mankin, A.S. | | Deposit date: | 2017-07-20 | | Release date: | 2018-02-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Co-produced natural ketolides methymycin and pikromycin inhibit bacterial growth by preventing synthesis of a limited number of proteins.
Nucleic Acids Res., 45, 2017
|
|
4Q9D
 
 | | X-ray structure of a putative thiamin diphosphate-dependent enzyme isolated from Mycobacterium smegmatis | | Descriptor: | Benzoylformate decarboxylase, FORMIC ACID, MAGNESIUM ION | | Authors: | Andrews, F.H, Horton, J.D, Yoon, H.J, Malik, A.M.K, Logsdon, M.G, Shin, D.H, Kneen, M.M, Suh, S.W, McLeish, M.J. | | Deposit date: | 2014-04-30 | | Release date: | 2015-04-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The kinetic characterization and X-ray structure of a putative benzoylformate decarboxylase from M. smegmatis highlights the difficulties in the functional annotation of ThDP-dependent enzymes.
Biochim.Biophys.Acta, 1854, 2015
|
|
3HDS
 
 | | Crystal structure of 4-methylmuconolactone methylisomerase in complex with MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-methylmuconolactone methylisomerase, short peptide ASWSA | | Authors: | Marin, M, Heinz, D.W, Pieper, D.H, Klink, B.U. | | Deposit date: | 2009-05-07 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure and catalytic mechanism of 4-methylmuconolactone methylisomerase
J.Biol.Chem., 284, 2009
|
|
2HYB
 
 | | Crystal Structure of Hexameric DsrEFH | | Descriptor: | DsrH, Intracellular sulfur oxidation protein dsrF, Putative sulfurtransferase dsrE | | Authors: | Shin, D.H, Connie, H, Schulte, A, Dahl, C, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2006-08-04 | | Release date: | 2007-07-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of hexameric DsrEFH
To be Published
|
|
2I15
 
 | |
2I1L
 
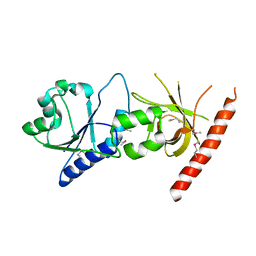 | | Crystal structure of the C2 form of FAD synthetase from Thermotoga maritima | | Descriptor: | Riboflavin kinase/FMN adenylyltransferase | | Authors: | Wang, W, Shin, D.H, Yokota, H, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2006-08-14 | | Release date: | 2006-11-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the C2 form of FAD synthetase from Thermotoga maritima
To be Published
|
|
4OIG
 
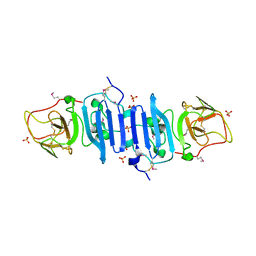 | |
3HK1
 
 | | Identification and Characterization of a Small Molecule Inhibitor of Fatty Acid Binding Proteins | | Descriptor: | 4-{[2-(methoxycarbonyl)-5-(2-thienyl)-3-thienyl]amino}-4-oxo-2-butenoic acid, Fatty acid-binding protein, adipocyte | | Authors: | Hertzel, A.V, Hellberg, K, Reynolds, J.M, Kruse, A.C, Juhlmann, B.E, Smith, A.J, Sanders, M.A, Ohlendorf, D.H, Suttles, J, Bernlohr, D.A. | | Deposit date: | 2009-05-22 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification and characterization of a small molecule inhibitor of Fatty Acid binding proteins.
J.Med.Chem., 52, 2009
|
|
2HY5
 
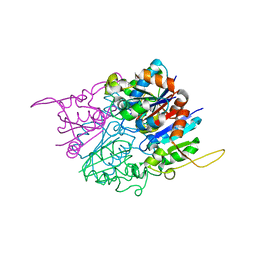 | | Crystal structure of DsrEFH | | Descriptor: | DsrH, Intracellular sulfur oxidation protein dsrF, Putative sulfurtransferase dsrE | | Authors: | Shin, D.H, Schulte, A, Dahl, C, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2006-08-04 | | Release date: | 2006-09-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structure of DsrEFH
To be Published
|
|
3HF5
 
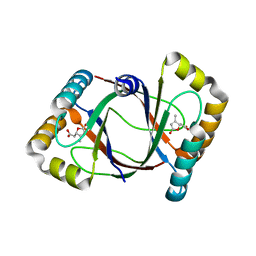 | | Crystal structure of 4-methylmuconolactone methylisomerase in complex with 3-methylmuconolactone | | Descriptor: | 4-methylmuconolactone methylisomerase, [(2S)-3-methyl-5-oxo-2,5-dihydrofuran-2-yl]acetic acid | | Authors: | Marin, M, Heinz, D.W, Pieper, D.H, Klink, B.U. | | Deposit date: | 2009-05-11 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure and catalytic mechanism of 4-methylmuconolactone methylisomerase
J.Biol.Chem., 284, 2009
|
|
3HFK
 
 | | Crystal structure of 4-methylmuconolactone methylisomerase (H52A) in complex with 4-methylmuconolactone | | Descriptor: | 4-methylmuconolactone methylisomerase, [(2S)-2-methyl-5-oxo-2,5-dihydrofuran-2-yl]acetic acid | | Authors: | Marin, M, Heinz, D.W, Pieper, D.H, Klink, B.U. | | Deposit date: | 2009-05-12 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and catalytic mechanism of 4-methylmuconolactone methylisomerase
J.Biol.Chem., 284, 2009
|
|
3F5H
 
 | | Crystal structure of fused docking domains from PikAIII and PikAIV of the pikromycin polyketide synthase | | Descriptor: | SODIUM ION, Type I polyketide synthase PikAIII, Type I polyketide synthase PikAIV fusion protein | | Authors: | Buchholz, T.J, Geders, T.W, Bartley, F.E, Reynolds, K.A, Smith, J.L, Sherman, D.H. | | Deposit date: | 2008-11-03 | | Release date: | 2009-01-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for binding specificity between subclasses of modular polyketide synthase docking domains.
Acs Chem.Biol., 4, 2009
|
|
2I1O
 
 | | Crystal Structure of a Nicotinate Phosphoribosyltransferase from Thermoplasma acidophilum | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ... | | Authors: | Shin, D.H, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2006-08-14 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of a zinc ion bound Nicotinate Phosphoribosyltransferase from Thermoplasma acidophilum
To be Published
|
|
2I14
 
 | |
4MQL
 
 | | Crystal structure of Antigen 85C-C209S mutant | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Diacylglycerol acyltransferase/mycolyltransferase Ag85C | | Authors: | Favrot, L, Grzegorzewicz, A.E, Lajiness, D.H, Marvin, R.K, Boucau, J, Isailovic, D, Jackson, M, Ronning, D.R. | | Deposit date: | 2013-09-16 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mechanism of inhibition of Mycobacterium tuberculosis antigen 85 by ebselen.
Nat Commun, 4, 2013
|
|
2GO9
 
 | | RRM domains 1 and 2 of Prp24 from S. cerevisiae | | Descriptor: | U4/U6 snRNA-associated splicing factor PRP24 | | Authors: | Reiter, N.J, Lee, D.H, Tonelli, M, Kwan, S.K, Brow, D.A, Butcher, S.E. | | Deposit date: | 2006-04-12 | | Release date: | 2007-02-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure and interactions of the first three RNA recognition motifs of splicing factor prp24.
J.Mol.Biol., 367, 2007
|
|
2GRL
 
 | | Crystal structure of dCT/iCF10 complex | | Descriptor: | PrgX, peptide | | Authors: | Shi, K, Kozlowicz, B.K, Gu, Z.Y, Ohlendorf, D.H, Earhart, C.A, Dunny, G.M. | | Deposit date: | 2006-04-24 | | Release date: | 2007-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular basis for control of conjugation by bacterial pheromone and inhibitor peptides.
Mol.Microbiol., 62, 2006
|
|
3IXX
 
 | | The pseudo-atomic structure of West Nile immature virus in complex with Fab fragments of the anti-fusion loop antibody E53 | | Descriptor: | E53 Fab Fragment (chain H), E53 Fab Fragment (chain L), Envelope protein E, ... | | Authors: | Cherrier, M.V, Kaufmann, B, Nybakken, G.E, Lok, S.M, Warren, J.T, Nelson, C.A, Kostyuchenko, V.A, Holdaway, H.A, Chipman, P.R, Kuhn, R.J, Diamond, M.S, Rossmann, M.G, Fremont, D.H. | | Deposit date: | 2009-02-26 | | Release date: | 2009-10-27 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | Structural basis for the preferential recognition of immature flaviviruses by a fusion-loop antibody
Embo J., 28, 2009
|
|
