1NTR
 
 | | SOLUTION STRUCTURE OF THE N-TERMINAL RECEIVER DOMAIN OF NTRC | | Descriptor: | NTRC RECEIVER DOMAIN | | Authors: | Volkman, B.F, Nohaile, M.J, Amy, N.K, Kustu, S, Wemmer, D.E. | | Deposit date: | 1994-09-16 | | Release date: | 1995-01-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the N-terminal receiver domain of NTRC.
Biochemistry, 34, 1995
|
|
1F08
 
 | | CRYSTAL STRUCTURE OF THE DNA-BINDING DOMAIN OF THE REPLICATION INITIATION PROTEIN E1 FROM PAPILLOMAVIRUS | | Descriptor: | BROMIDE ION, REPLICATION PROTEIN E1 | | Authors: | Enemark, E.J, Chen, G, Vaughn, D.E, Stenlund, A, Joshua-Tor, L. | | Deposit date: | 2000-05-15 | | Release date: | 2001-05-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the DNA binding domain of the replication initiation protein E1 from papillomavirus.
Mol.Cell, 6, 2000
|
|
1P8Q
 
 | | Structural and Functional Importance of First-Shell Metal Ligands in the Binuclear Cluster of Arginase I. | | Descriptor: | Arginase 1, GLYCEROL, MANGANESE (II) ION | | Authors: | Cama, E, Emig, F.A, Ash, D.E, Christianson, D.W. | | Deposit date: | 2003-05-07 | | Release date: | 2003-06-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural and functional importance of first-shell metal ligands in the binuclear
manganese cluster of arginase I
Biochemistry, 42, 2003
|
|
1F4H
 
 | | E. COLI (LACZ) BETA-GALACTOSIDASE (ORTHORHOMBIC) | | Descriptor: | BETA-GALACTOSIDASE, MAGNESIUM ION | | Authors: | Juers, D.H, Jacobson, R.H, Wigley, D, Zhang, X.J, Huber, R.E, Tronrud, D.E, Matthews, B.W. | | Deposit date: | 2000-06-07 | | Release date: | 2001-02-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | High resolution refinement of beta-galactosidase in a new crystal form reveals multiple metal-binding sites and provides a structural basis for alpha-complementation.
Protein Sci., 9, 2000
|
|
1F4A
 
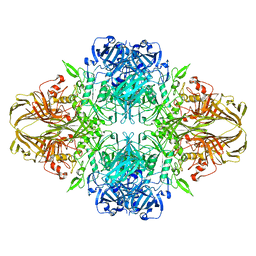 | | E. COLI (LACZ) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-ORTHORHOMBIC) | | Descriptor: | BETA-GALACTOSIDASE, MAGNESIUM ION | | Authors: | Juers, D.H, Jacobson, R.H, Wigley, D, Zhang, X.J, Huber, R.E, Tronrud, D.E, Matthews, B.W. | | Deposit date: | 2000-06-07 | | Release date: | 2001-02-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | High resolution refinement of beta-galactosidase in a new crystal form reveals multiple metal-binding sites and provides a structural basis for alpha-complementation.
Protein Sci., 9, 2000
|
|
1DUT
 
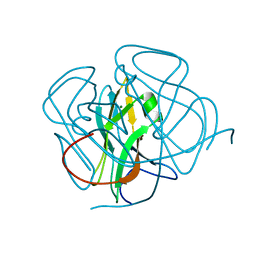 | | FIV DUTP PYROPHOSPHATASE | | Descriptor: | DUTP PYROPHOSPHATASE, MAGNESIUM ION | | Authors: | Prasad, G.S, Stura, E.A, Mcree, D.E, Laco, G.S, Hasselkus-Light, C, Elder, J.H, Stout, C.D. | | Deposit date: | 1996-09-15 | | Release date: | 1997-01-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of dUTP pyrophosphatase from feline immunodeficiency virus.
Protein Sci., 5, 1996
|
|
1P8F
 
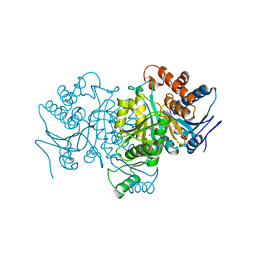 | |
1DT1
 
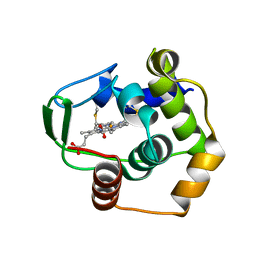 | | THERMUS THERMOPHILUS CYTOCHROME C552 SYNTHESIZED BY ESCHERICHIA COLI | | Descriptor: | CYTOCHROME C552, HEME C | | Authors: | Fee, J.A, Chen, Y, Hill, M.J, Gomez-Moran, E, Loehr, T, Ai, J, Thony-Meyer, L, Williams, P.A, Stura, E, Sridhar, V, McRee, D.E. | | Deposit date: | 2000-01-10 | | Release date: | 2000-02-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Integrity of thermus thermophilus cytochrome c552 synthesized by Escherichia coli cells expressing the host-specific cytochrome c maturation genes, ccmABCDEFGH: biochemical, spectral, and structural characterization of the recombinant protein.
Protein Sci., 9, 2000
|
|
1DSP
 
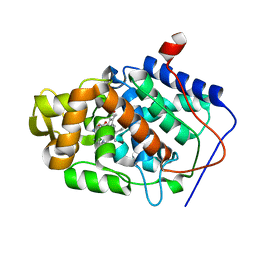 | | CYTOCHROME C PEROXIDASE H175G MUTANT, IMIDAZOLE COMPLEX AT PH 7, ROOM TEMPERATURE. | | Descriptor: | CYTOCHROME C PEROXIDASE, IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hirst, J, Wilcox, S.K, Williams, P.A, McRee, D.E, Goodin, D.B. | | Deposit date: | 2000-01-07 | | Release date: | 2001-03-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Replacement of the axial histidine ligand with imidazole in cytochrome c peroxidase. 1. Effects on structure.
Biochemistry, 40, 2001
|
|
1NPZ
 
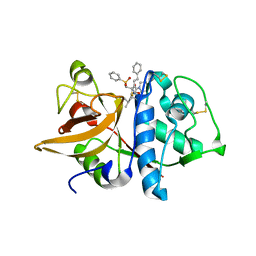 | | Crystal structures of Cathepsin S inhibitor complexes | | Descriptor: | Cathepsin S, N~2~-(morpholin-4-ylcarbonyl)-N-[(3S)-1-phenyl-5-(phenylsulfonyl)pentan-3-yl]-L-leucinamide | | Authors: | Pauly, T.A, Sulea, T, Ammirati, M, Sivaraman, J, Danley, D.E, Griffor, M.C, Kamath, A.V, Wang, I.K, Laird, E.R, Seddon, A.P, Menard, R, Cygler, M, Rath, V.L. | | Deposit date: | 2003-01-20 | | Release date: | 2003-04-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Specificity determinants of human cathepsin s revealed
by crystal structures of complexes.
Biochemistry, 42, 2003
|
|
1NNF
 
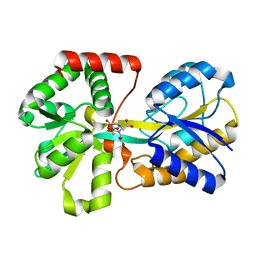 | | Crystal Structure Analysis of Haemophlius Influenzae Ferric-ion Binding Protein H9Q Mutant Form | | Descriptor: | FE (III) ION, Iron-utilization periplasmic protein, {[-(BIS-CARBOXYMETHYL-AMINO)-ETHYL]-CARBOXYMETHYL-AMINO}-ACETIC ACID | | Authors: | Shouldice, S.R, Dougan, D.R, Skene, R.J, Tari, L.W, McRee, D.E, Yu, R.-H, Schryvers, A.B. | | Deposit date: | 2003-01-13 | | Release date: | 2003-04-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | High Resolution Structure of an Alternate Form of the Ferric ion Binding Protein from Haemophilus influenzae
J.Biol.Chem., 278, 2003
|
|
1DSG
 
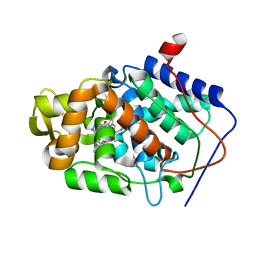 | | CYTOCHROME C PEROXIDASE H175G MUTANT, IMIDAZOLE COMPLEX AT PH 5, ROOM TEMPERATURE. | | Descriptor: | CYTOCHROME C PEROXIDASE, IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hirst, J, Wilcox, S.K, Williams, P.A, McRee, D.E, Goodin, D.B. | | Deposit date: | 2000-01-07 | | Release date: | 2001-03-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Replacement of the axial histidine ligand with imidazole in cytochrome c peroxidase. 1. Effects on structure.
Biochemistry, 40, 2001
|
|
1DZ9
 
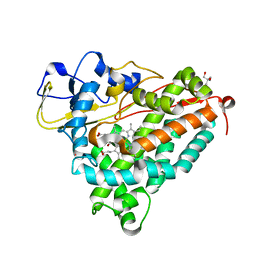 | | Putative oxo complex of P450cam from Pseudomonas putida | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CAMPHOR, CYTOCHROME P450-CAM, ... | | Authors: | Schlichting, I, Berendzen, J, Chu, K, Stock, A.M, Maves, S.A, Benson, D.E, Sweet, R.M, Ringe, D, Petsko, G.A, Sligar, S.G. | | Deposit date: | 2000-02-18 | | Release date: | 2000-03-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Catalytic Pathway of Cytochrome P450Cam at Atomic Resolution
Science, 287, 2000
|
|
1DZ4
 
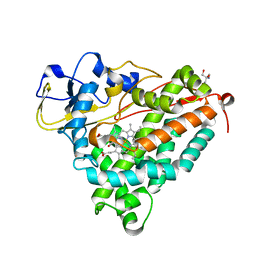 | | ferric p450cam from pseudomonas putida | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CAMPHOR, CYTOCHROME P450-CAM, ... | | Authors: | Schlichting, I, Berendzen, J, Chu, K, Stock, A.M, Maves, S.A, Benson, D.E, Sweet, R.M, Ringe, D, Petsko, G.A, Sligar, S.G. | | Deposit date: | 2000-02-16 | | Release date: | 2000-07-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Catalytic Pathway of Cytochrome P450Cam at Atomic Resolution
Science, 287, 2000
|
|
1DZ6
 
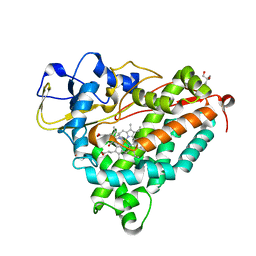 | | ferrous p450cam from pseudomonas putida | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CAMPHOR, CYTOCHROME P450-CAM, ... | | Authors: | Schlichting, I, Berendzen, J, Chu, K, Stock, A.M, Maves, S.A, Benson, D.E, Sweet, R.M, Ringe, D, Petsko, G.A, Sligar, S.G. | | Deposit date: | 2000-02-18 | | Release date: | 2000-03-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Catalytic Pathway of Cytochrome P450Cam at Atomic Resolution
Science, 287, 2000
|
|
1DZ8
 
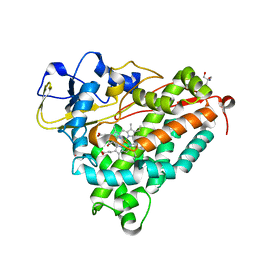 | | oxygen complex of p450cam from pseudomonas putida | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CAMPHOR, CYTOCHROME P450-CAM, ... | | Authors: | Schlichting, I, Berendzen, J, Chu, K, Stock, A.M, Maves, S.A, Benson, D.E, Sweet, R.M, Ringe, D, Petsko, G.A, Sligar, S.G. | | Deposit date: | 2000-02-18 | | Release date: | 2000-03-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Catalytic Pathway of Cytochrome P450Cam at Atomic Resolution
Science, 287, 2000
|
|
1MZA
 
 | | crystal structure of human pro-granzyme K | | Descriptor: | pro-granzyme K | | Authors: | Hink-Schauer, C, Estebanez-Perpina, E, Wilharm, E, Fuentes-Prior, P, Klinkert, W, Bode, W, Jenne, D.E. | | Deposit date: | 2002-10-07 | | Release date: | 2003-01-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | The 2.2-A Crystal Structure of Human Pro-granzyme K Reveals a Rigid Zymogen with Unusual Features
J.BIOL.CHEM., 277, 2002
|
|
1FOC
 
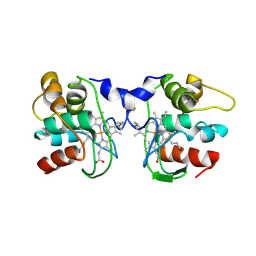 | | Cytochrome C557: improperly folded thermus thermophilus C552 | | Descriptor: | CYTOCHROME RC557, HEME C | | Authors: | McRee, D.E, Williams, P.A, Fee, J.A, Bren, K.L. | | Deposit date: | 2000-08-27 | | Release date: | 2000-11-08 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Recombinant cytochrome rC557 obtained from Escherichia coli cells expressing a truncated Thermus thermophilus cycA gene. Heme inversion in an improperly matured protein
J.Biol.Chem., 276, 2001
|
|
1FC0
 
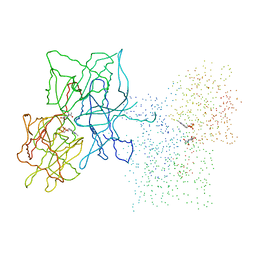 | | HUMAN LIVER GLYCOGEN PHOSPHORYLASE COMPLEXED WITH N-ACETYL-BETA-D-GLUCOPYRANOSYLAMINE | | Descriptor: | GLYCOGEN PHOSPHORYLASE, LIVER FORM, N-acetyl-beta-D-glucopyranosylamine, ... | | Authors: | Rath, V.L, Ammirati, M, LeMotte, P.K, Fennell, K.F, Mansour, M.M, Danley, D.E, Hynes, T.R, Schulte, G.K, Wasilko, D.J, Pandit, J. | | Deposit date: | 2000-07-17 | | Release date: | 2000-08-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Activation of human liver glycogen phosphorylase by alteration of the secondary structure and packing of the catalytic core.
Mol.Cell, 6, 2000
|
|
1FJ7
 
 | |
1FEO
 
 | |
1O7Y
 
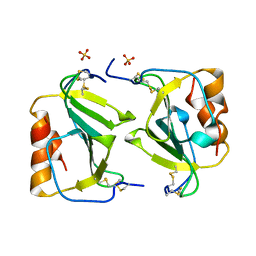 | | Crystal structure of IP-10 M-form | | Descriptor: | SMALL INDUCIBLE CYTOKINE B10, SULFATE ION | | Authors: | Swaminathan, G.J, Holloway, D.E, Papageorgiou, A.C, Acharya, K.R. | | Deposit date: | 2002-11-20 | | Release date: | 2003-05-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structures of Oligomeric Forms of the Ip-10/Cxcl10 Chemokine
Structure, 11, 2003
|
|
1P77
 
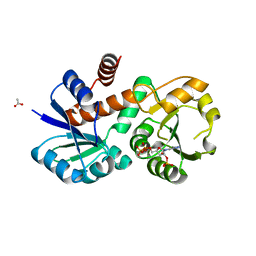 | | CRYSTAL STRUCTURE OF SHIKIMATE DEHYDROGENASE (AROE) FROM HAEMOPHILUS INFLUENZAE | | Descriptor: | 2'-MONOPHOSPHOADENOSINE-5'-DIPHOSPHATE, ACETATE ION, Shikimate 5-dehydrogenase | | Authors: | Ye, S, von Delft, F, Brooun, A, Knuth, M.W, Swanson, R.V, McRee, D.E. | | Deposit date: | 2003-04-30 | | Release date: | 2003-08-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The crystal structure of shikimate dehydrogenase (AroE) reveals a unique NADPH binding mode
J.Bacteriol., 185, 2003
|
|
1P8N
 
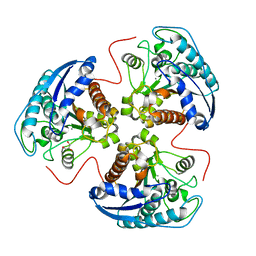 | | Structural and Functional Importance of First-Shell Metal Ligands in the Binuclear Manganese Cluster of Arginase I. | | Descriptor: | Arginase 1, GLYCEROL, MANGANESE (II) ION | | Authors: | Cama, E, Emig, F.A, Ash, D.E, Christianson, D.W. | | Deposit date: | 2003-05-07 | | Release date: | 2003-06-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and functional importance of first-shell metal ligands in the binuclear
manganese cluster of arginase I
Biochemistry, 42, 2003
|
|
1P31
 
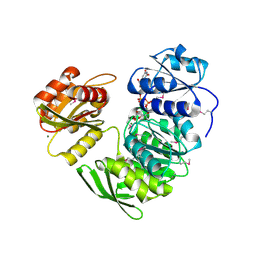 | | Crystal Structure of UDP-N-acetylmuramic acid:L-alanine Ligase (MurC) from Haemophilus influenzae | | Descriptor: | MAGNESIUM ION, UDP-N-acetylmuramate--alanine ligase, URIDINE-DIPHOSPHATE-2(N-ACETYLGLUCOSAMINYL) BUTYRIC ACID | | Authors: | Mol, C.D, Brooun, A, Dougan, D.R, Hilgers, M.T, Tari, L.W, Wijnands, R.A, Knuth, M.W, McRee, D.E, Swanson, R.V. | | Deposit date: | 2003-04-16 | | Release date: | 2003-07-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structures of Active Fully Assembled Substrate- and Product-Bound Complexes of UDP-N-Acetylmuramic Acid:L-Alanine Ligase (MurC) from Haemophilus influenzae.
J.Bacteriol., 185, 2003
|
|
