6BFD
 
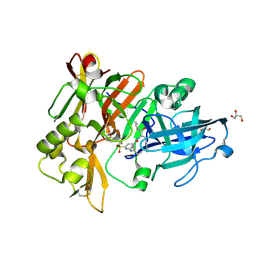 | | BACE crystal structure with hydroxy pyrrolidine inhibitor | | Descriptor: | 2-{[(2S)-butan-2-yl]amino}-N-{(1R,2S)-1-hydroxy-3-phenyl-1-[(2R)-pyrrolidin-2-yl]propan-2-yl}-6-(methylsulfonyl)pyridine-4-carboxamide, Beta-secretase 1, GLYCEROL | | Authors: | Timm, D.E. | | Deposit date: | 2017-10-26 | | Release date: | 2017-11-15 | | Last modified: | 2017-12-27 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Optimization of Hydroxyethylamine Transition State Isosteres as Aspartic Protease Inhibitors by Exploiting Conformational Preferences.
J. Med. Chem., 60, 2017
|
|
6BFW
 
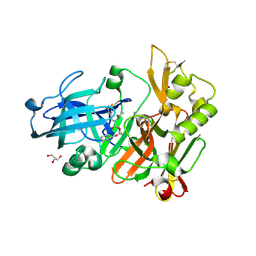 | | BACE crystal structure with hydroxy morpholine inhibitor | | Descriptor: | Beta-secretase 1, GLYCEROL, N-[(1S,2S)-1-[(3R,6R)-6-(cyclohexylmethoxy)morpholin-3-yl]-3-(3,5-difluorophenyl)-1-hydroxypropan-2-yl]acetamide | | Authors: | Timm, D.E. | | Deposit date: | 2017-10-27 | | Release date: | 2017-11-15 | | Last modified: | 2017-12-27 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Optimization of Hydroxyethylamine Transition State Isosteres as Aspartic Protease Inhibitors by Exploiting Conformational Preferences.
J. Med. Chem., 60, 2017
|
|
1S6N
 
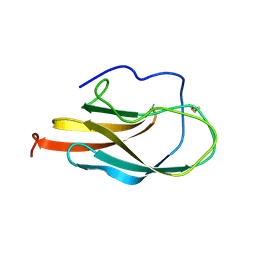 | | NMR Structure of Domain III of the West Nile Virus Envelope Protein, Strain 385-99 | | Descriptor: | envelope glycoprotein | | Authors: | Volk, D.E, Beasley, D.W, Kallick, D.A, Holbrook, M.R, Barrett, A.D, Gorenstein, D.G. | | Deposit date: | 2004-01-26 | | Release date: | 2004-07-06 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Antibody Binding Studies of the Envelope Protein Domain III from the New York Strain of West Nile Virus
J.Biol.Chem., 279, 2004
|
|
4NHP
 
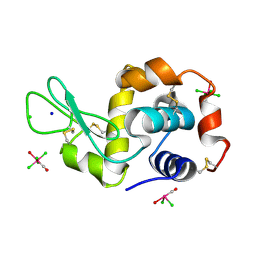 | | X-ray structure of the complex between the hen egg white lysozyme and pentachlorocarbonyliridate (III) (4 days) | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION, ... | | Authors: | Petruk, A.A, Bikiel, D.E, Vergara, A, Merlino, A. | | Deposit date: | 2013-11-05 | | Release date: | 2014-09-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Interaction between proteins and Ir based CO releasing molecules: mechanism of adduct formation and CO release.
Inorg.Chem., 53, 2014
|
|
4NHT
 
 | | X-ray structure of the complex between hen egg white lysozyme and pentachlorocarbonyliridate(III) (6 days) | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION, ... | | Authors: | Petruk, A.A, Bikiel, D.E, Vergara, A, Merlino, A. | | Deposit date: | 2013-11-05 | | Release date: | 2014-09-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Interaction between proteins and Ir based CO releasing molecules: mechanism of adduct formation and CO release.
Inorg.Chem., 53, 2014
|
|
4NHS
 
 | | X-ray structure of the complex between hen egg white lysozyme and pentachlorocarbonyliridate(III) (9 days) | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION, ... | | Authors: | Petruk, A.A, Bikiel, D.E, Vergara, A, Merlino, A. | | Deposit date: | 2013-11-05 | | Release date: | 2014-09-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Interaction between proteins and Ir based CO releasing molecules: mechanism of adduct formation and CO release.
Inorg.Chem., 53, 2014
|
|
4NIJ
 
 | | X-ray structure of the complex between hen egg white lysozyme and pentachlorocarbonyliridate(III) (30 days) | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION, ... | | Authors: | Petruk, A.A, Bikiel, D.E, Vergara, A, Merlino, A. | | Deposit date: | 2013-11-06 | | Release date: | 2014-09-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Interaction between proteins and Ir based CO releasing molecules: mechanism of adduct formation and CO release.
Inorg.Chem., 53, 2014
|
|
4NHQ
 
 | | X-ray structure of the complex between hen egg white lysozyme and pentachlorocarbonyliridate(III) (5 days) | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION, ... | | Authors: | Petruk, A.A, Bikiel, D.E, Vergara, A, Merlino, A. | | Deposit date: | 2013-11-05 | | Release date: | 2014-09-17 | | Last modified: | 2015-06-24 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Interaction between proteins and Ir based CO releasing molecules: mechanism of adduct formation and CO release.
Inorg.Chem., 53, 2014
|
|
4N9R
 
 | | X-ray structure of the complex between hen egg white lysozyme and pentacholrocarbonyliridate(III) (1 day) | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION, ... | | Authors: | Petruk, A.A, Bikiel, D.E, Vergara, A, Merlino, A. | | Deposit date: | 2013-10-21 | | Release date: | 2015-12-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Interaction between proteins and Ir based CO releasing molecules: mechanism of adduct formation and CO release.
Inorg.Chem., 53, 2014
|
|
5IH9
 
 | | MELK in complex with NVS-MELK8A | | Descriptor: | 1-methyl-4-[4-(4-{3-[(piperidin-4-yl)methoxy]pyridin-4-yl}-1H-pyrazol-1-yl)phenyl]piperazine, Maternal embryonic leucine zipper kinase | | Authors: | Sprague, E.R, Puleo, D.E. | | Deposit date: | 2016-02-29 | | Release date: | 2016-06-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Toward the Validation of Maternal Embryonic Leucine Zipper Kinase: Discovery, Optimization of Highly Potent and Selective Inhibitors, and Preliminary Biology Insight.
J.Med.Chem., 59, 2016
|
|
5IH8
 
 | | MELK in complex with NVS-MELK1 | | Descriptor: | Maternal embryonic leucine zipper kinase, N-[(pyridin-2-yl)methyl]-4-[4-(pyridin-4-yl)-1H-pyrazol-1-yl]benzamide | | Authors: | Sprague, E.R, Puleo, D.E. | | Deposit date: | 2016-02-29 | | Release date: | 2016-06-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Toward the Validation of Maternal Embryonic Leucine Zipper Kinase: Discovery, Optimization of Highly Potent and Selective Inhibitors, and Preliminary Biology Insight.
J.Med.Chem., 59, 2016
|
|
2QS1
 
 | | Crystal structure of the GluR5 ligand binding core dimer in complex with UBP315 at 1.80 Angstroms resolution | | Descriptor: | 3-({3-[(2S)-2-amino-2-carboxyethyl]-5-methyl-2,6-dioxo-3,6-dihydropyrimidin-1(2H)-yl}methyl)-4,5-dibromothiophene-2-carboxylic acid, CHLORIDE ION, Glutamate receptor, ... | | Authors: | Alushin, G.M, Jane, D.E, Mayer, M.L. | | Deposit date: | 2007-07-30 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Binding site and ligand flexibility revealed by high resolution crystal structures of GluK1 competitive antagonists.
Neuropharmacology, 60, 2011
|
|
2QS2
 
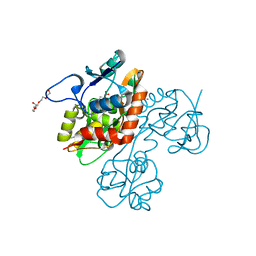 | | Crystal structure of the GluR5 ligand binding core dimer in complex with UBP318 at 1.80 Angstroms resolution | | Descriptor: | 3-({3-[(2S)-2-amino-2-carboxyethyl]-5-bromo-2,6-dioxo-3,6-dihydropyrimidin-1(2H)-yl}methyl)thiophene-2-carboxylic acid, CHLORIDE ION, Glutamate receptor, ... | | Authors: | Alushin, G.M, Jane, D.E, Mayer, M.L. | | Deposit date: | 2007-07-30 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Binding site and ligand flexibility revealed by high resolution crystal structures of GluK1 competitive antagonists.
Neuropharmacology, 60, 2011
|
|
2QS4
 
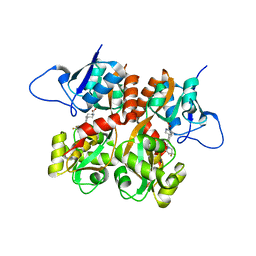 | | Crystal structure of the GluR5 ligand binding core dimer in complex with LY466195 at 1.58 Angstroms resolution | | Descriptor: | (3S,4aR,6S,8aR)-6-{[(2S)-2-carboxy-4,4-difluoropyrrolidin-1-yl]methyl}decahydroisoquinoline-3-carboxylic acid, AMMONIUM ION, GLYCEROL, ... | | Authors: | Alushin, G.M, Jane, D.E, Mayer, M.L. | | Deposit date: | 2007-07-30 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Binding site and ligand flexibility revealed by high resolution crystal structures of GluK1 competitive antagonists.
Neuropharmacology, 60, 2011
|
|
4FS3
 
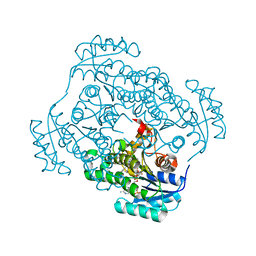 | | Crystal structure of Staphylococcus aureus enoyl-ACP reductase in complex with NADP and AFN-1252 | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADPH] FabI, N-methyl-N-[(3-methyl-1-benzofuran-2-yl)methyl]-3-(7-oxo-5,6,7,8-tetrahydro-1,8-naphthyridin-3-yl)propanamide, [[(2R,3S,4R,5R)-5-(3-aminocarbonyl-4H-pyridin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Kaplan, N, Yethon, J, Bardouniotis, E, Thalakada, R, Albert, M, Awrey, D.E, Romanov, V, Dorsey, M, Ramnauth, J, Clarke, T.E, Schmid, M.B, Berman, J, Pauls, H.W. | | Deposit date: | 2012-06-26 | | Release date: | 2012-09-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mode of Action, In Vitro Activity, and In Vivo Efficacy of AFN-1252, a Selective Antistaphylococcal FabI Inhibitor.
Antimicrob.Agents Chemother., 56, 2012
|
|
1ZUQ
 
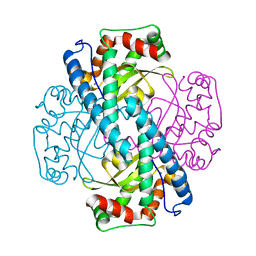 | | Contribution to Structure and Catalysis of Tyrosine 34 in Human Manganese Superoxide Dismutase | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase | | Authors: | Hearn, A.S, Perry, J.J, Cabelii, D.E, Tainer, J.A, Nick, H.S, Silverman, D.S. | | Deposit date: | 2005-05-31 | | Release date: | 2006-05-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Contribution of human manganese superoxide dismutase tyrosine 34 to structure and catalysis.
Biochemistry, 48, 2009
|
|
1ZTE
 
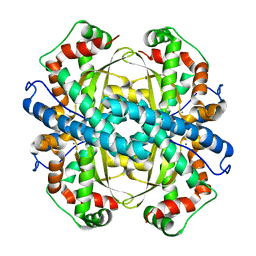 | | Contribution to Structure and Catalysis of Tyrosine 34 in Human Manganese Suerpoxide Dismutase | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase [Mn], mitochondrial | | Authors: | Hearn, A.S, Perry, J.J, Cabelii, D.E, Tainer, J.A, Nick, H.S, Silverman, D.S. | | Deposit date: | 2005-05-26 | | Release date: | 2006-05-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Contribution of human manganese superoxide dismutase tyrosine 34 to structure and catalysis.
Biochemistry, 48, 2009
|
|
2CUA
 
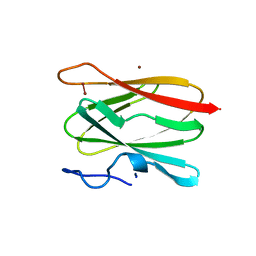 | | THE CUA DOMAIN OF CYTOCHROME BA3 FROM THERMUS THERMOPHILUS | | Descriptor: | DINUCLEAR COPPER ION, PROTEIN (CUA), ZINC ION | | Authors: | Williams, P.A, Blackburn, N.J, Sanders, D, Bellamy, H, Stura, E.A, Fee, J.A, Mcree, D.E. | | Deposit date: | 1999-02-18 | | Release date: | 1999-05-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The CuA domain of Thermus thermophilus ba3-type cytochrome c oxidase at 1.6 A resolution.
Nat.Struct.Biol., 6, 1999
|
|
1OYE
 
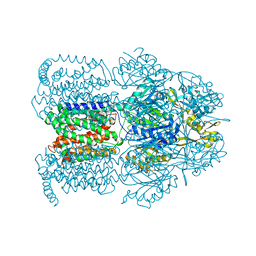 | | Structural Basis of Multiple Binding Capacity of the AcrB multidrug Efflux Pump | | Descriptor: | 1-CYCLOPROPYL-6-FLUORO-4-OXO-7-PIPERAZIN-1-YL-1,4-DIHYDROQUINOLINE-3-CARBOXYLIC ACID, Acriflavine resistance protein B | | Authors: | Yu, E.W, McDermott, G, Zgurskaya, H.I, Nikaido, H, Koshland Jr, D.E. | | Deposit date: | 2003-04-03 | | Release date: | 2003-05-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.48 Å) | | Cite: | Structural basis of multiple drug-binding capacity of the AcrB multidrug efflux pump.
Science, 300, 2003
|
|
2DCX
 
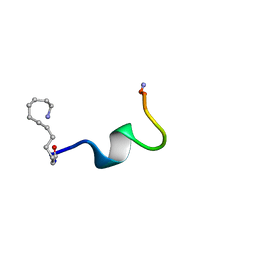 | | NMR solution structure of the Dermaseptin antimicrobial peptide analog NC12-K4S4(1-13)a | | Descriptor: | 12-AMINO-DODECANOIC ACID, Dermaseptin-4 | | Authors: | Shalev, D.E, Rotem, S, Fish, A, Mor, A. | | Deposit date: | 2006-01-17 | | Release date: | 2006-02-28 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Consequences of N-acylation on structure and membrane binding properties of dermaseptin derivative k4-s4-(1-13)
J.Biol.Chem., 281, 2006
|
|
2DD6
 
 | | Solution structure of Dermaseptin antimicrobial peptide truncated, mutated analog, K4-S4(1-13)a | | Descriptor: | Dermaseptin-4 | | Authors: | Shalev, D.E, Rotem, S, Fish, A, Mor, A. | | Deposit date: | 2006-01-19 | | Release date: | 2006-02-28 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Consequences of N-acylation on structure and membrane binding properties of dermaseptin derivative k4-s4-(1-13)
J.Biol.Chem., 281, 2006
|
|
1ECI
 
 | | ECTATOMIN (WATER SOLUTION, NMR 20 STRUCTURES) | | Descriptor: | ECTATOMIN | | Authors: | Nolde, D.E, Sobol, A.G, Pluzhnikov, K.A, Arseniev, A.S, Grishin, E.V. | | Deposit date: | 1995-08-16 | | Release date: | 1995-12-07 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of ectatomin from Ectatomma tuberculatum ant venom.
J.Biomol.NMR, 5, 1995
|
|
2BXR
 
 | | Human Monoamine Oxidase A in complex with Clorgyline, Crystal Form A | | Descriptor: | AMINE OXIDASE [FLAVIN-CONTAINING] A, FLAVIN-ADENINE DINUCLEOTIDE, N-[3-(2,4-DICHLOROPHENOXY)PROPYL]-N-METHYL-N-PROP-2-YNYLAMINE | | Authors: | De Colibus, L, Binda, C, Edmondson, D.E, Mattevi, A. | | Deposit date: | 2005-07-27 | | Release date: | 2005-08-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Three-Dimensional Structure of Human Monoamine Oxidase a (Mao A): Relation to the Structures of Rat Mao a and Human Mao B
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1E74
 
 | | NMR SOLUTION STRUCTURE OF ALPHA-CONOTOXIN IM1 POINT MUTATION VARIANT R11E | | Descriptor: | ALPHA-CONOTOXIN IM1(R11E) | | Authors: | Rogers, J.P, Luginbuhl, P, Pemberton, K, Harty, P, Wemmer, D.E, Stevens, R.C. | | Deposit date: | 2000-08-24 | | Release date: | 2000-12-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure-Activity Relationships in a Peptidic Alpha7 Nicotinic Acetylcholine Receptor Antagonist
J.Mol.Biol., 304, 2000
|
|
1E76
 
 | | NMR SOLUTION STRUCTURE OF ALPHA-CONOTOXIN IM1 POINT MUTATION VARIANT D5N | | Descriptor: | ALPHA-CONOTOXIN IM1(D5N) | | Authors: | Rogers, J.P, Luginbuhl, P, Pemberton, K, Harty, P, Wemmer, D.E, Stevens, R.C. | | Deposit date: | 2000-08-24 | | Release date: | 2000-12-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure-Activity Relationships in a Peptidic Alpha7 Nicotinic Acetylcholine Receptor Antagonist
J.Mol.Biol., 304, 2000
|
|
