2ITF
 
 | | Crystal structure IsdA NEAT domain from Staphylococcus aureus with heme bound | | Descriptor: | Iron-regulated surface determinant protein A, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Grigg, J.C, Vermeiren, C.L, Heinrichs, D.E, Murphy, M.E. | | Deposit date: | 2006-10-19 | | Release date: | 2006-12-26 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Haem recognition by a Staphylococcus aureus NEAT domain.
Mol.Microbiol., 63, 2007
|
|
2ITE
 
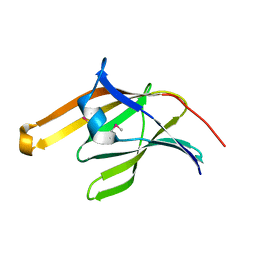 | | Crystal structure of the IsdA NEAT domain from Staphylococcus aureus | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Iron-regulated surface determinant protein A | | Authors: | Grigg, J.C, Vermeiren, C.L, Heinrichs, D.E, Murphy, M.E. | | Deposit date: | 2006-10-19 | | Release date: | 2006-12-26 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Haem recognition by a Staphylococcus aureus NEAT domain.
Mol.Microbiol., 63, 2007
|
|
6CTB
 
 | |
6D74
 
 | |
6DU8
 
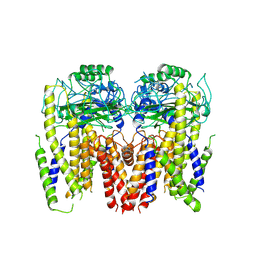 | | Human Polycsytin 2-l1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Polycystic kidney disease 2-like 1 protein | | Authors: | Hulse, R.E, Clapham, D.E, Li, Z, Huang, R.K, Zhang, J. | | Deposit date: | 2018-06-20 | | Release date: | 2018-07-25 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Cryo-EM structure of the polycystin 2-l1 ion channel.
Elife, 7, 2018
|
|
2J4T
 
 | | Biological and Structural Features of Murine Angiogenin-4, an Angiogenic Protein | | Descriptor: | ANGIOGENIN-4 | | Authors: | Crabtree, B, Holloway, D.E, Baker, M.D, Acharya, K.R, Subramanian, V. | | Deposit date: | 2006-09-05 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Biological and Structural Features of Murine Angiogenin-4, an Angiogenic Protein
Biochemistry, 46, 2007
|
|
7KIO
 
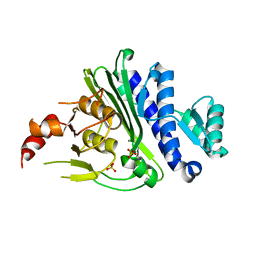 | | Crystal structure of inositol polyphosphate 1-phosphatase (INPP1) D54A mutant | | Descriptor: | CALCIUM ION, Inositol polyphosphate 1-phosphatase, SULFATE ION | | Authors: | Xiong, J.-P, Dollins, D.E, Ren, Y, York, J.D. | | Deposit date: | 2020-10-24 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A structural basis for lithium and substrate binding of an inositide phosphatase.
J.Biol.Chem., 296, 2020
|
|
1IDE
 
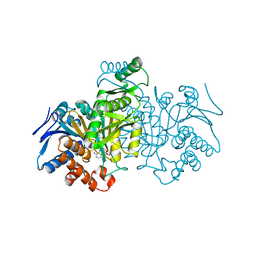 | | ISOCITRATE DEHYDROGENASE Y160F MUTANT STEADY-STATE INTERMEDIATE COMPLEX (LAUE DETERMINATION) | | Descriptor: | ISOCITRATE DEHYDROGENASE, ISOCITRIC ACID, MAGNESIUM ION, ... | | Authors: | Bolduc, J.M, Dyer, D.H, Scott, W.G, Singer, P, Sweet, R.M, Koshland Junior, D.E, Stoddard, B.L. | | Deposit date: | 1995-01-18 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mutagenesis and Laue structures of enzyme intermediates: isocitrate dehydrogenase.
Science, 268, 1995
|
|
7KIR
 
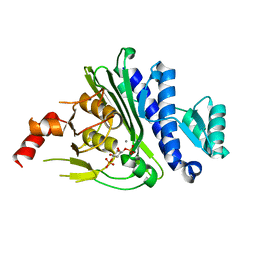 | | Crystal structure of inositol polyphosphate 1-phosphatase (INPP1) D54A mutant in complex with inositol (1,4)-bisphosphate | | Descriptor: | CALCIUM ION, D-MYO-INOSITOL-1,4-BISPHOSPHATE, Inositol polyphosphate 1-phosphatase | | Authors: | Dollins, D.E, Xiong, J.-P, Ren, Y, York, J.D. | | Deposit date: | 2020-10-24 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A structural basis for lithium and substrate binding of an inositide phosphatase.
J.Biol.Chem., 296, 2020
|
|
1IDC
 
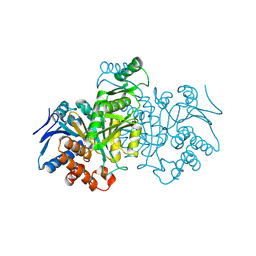 | | ISOCITRATE DEHYDROGENASE FROM E.COLI (MUTANT K230M), STEADY-STATE INTERMEDIATE COMPLEX DETERMINED BY LAUE CRYSTALLOGRAPHY | | Descriptor: | 2-OXALOSUCCINIC ACID, ISOCITRATE DEHYDROGENASE, MAGNESIUM ION | | Authors: | Bolduc, J.M, Dyer, D.H, Scott, W.G, Singer, P, Sweet, R.M, Koshland Junior, D.E, Stoddard, B.L. | | Deposit date: | 1995-01-18 | | Release date: | 1996-03-08 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mutagenesis and Laue structures of enzyme intermediates: isocitrate dehydrogenase.
Science, 268, 1995
|
|
1IDF
 
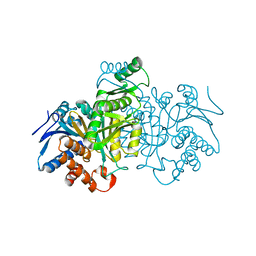 | | ISOCITRATE DEHYDROGENASE K230M MUTANT APO ENZYME | | Descriptor: | ISOCITRATE DEHYDROGENASE | | Authors: | Bolduc, J.M, Dyer, D.H, Scott, W.G, Singer, P, Sweet, R.M, Koshland Junior, D.E, Stoddard, B.L. | | Deposit date: | 1995-01-18 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mutagenesis and Laue structures of enzyme intermediates: isocitrate dehydrogenase.
Science, 268, 1995
|
|
2VRL
 
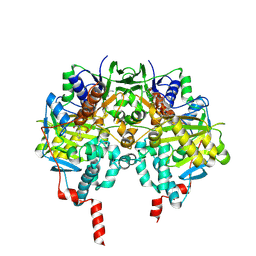 | | Structure of human MAO B in complex with benzylhydrazine | | Descriptor: | AMINE OXIDASE [FLAVIN-CONTAINING] B, FLAVIN-ADENINE DINUCLEOTIDE, TOLUENE | | Authors: | Binda, C, Wang, J, Li, M, Hubalek, F, Mattevi, A, Edmondson, D.E. | | Deposit date: | 2008-04-09 | | Release date: | 2008-04-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Mechanistic Studies of Arylalkylhydrazine Inhibition of Human Monoamine Oxidases a and B
Biochemistry, 47, 2008
|
|
2VRM
 
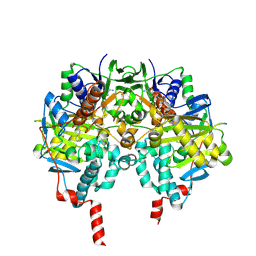 | | Structure of human MAO B in complex with phenyethylhydrazine | | Descriptor: | AMINE OXIDASE [FLAVIN-CONTAINING] B, FLAVIN-ADENINE DINUCLEOTIDE, PHENYLETHANE | | Authors: | Binda, C, Wang, J, Li, M, Hubalek, F, Mattevi, A, Edmondson, D.E. | | Deposit date: | 2008-04-09 | | Release date: | 2008-04-22 | | Last modified: | 2014-01-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Mechanistic Studies of Arylalkylhydrazine Inhibition of Human Monoamine Oxidases a and B
Biochemistry, 47, 2008
|
|
2W5G
 
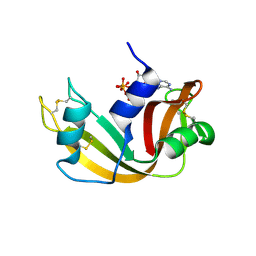 | | RNASE A-5'-ATP COMPLEX | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, RIBONUCLEASE PANCREATIC | | Authors: | Chavali, G.B, Holloway, D.E, Baker, M.D, Acharya, K.R. | | Deposit date: | 2008-12-10 | | Release date: | 2009-02-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Influence of Naturally-Occurring 5'-Pyrophosphate- Linked Substituents on the Binding of Adenylic Inhibitors to Ribonuclease A: An X-Ray Crystallographic Study.
Biopolymers, 91, 2009
|
|
2W5L
 
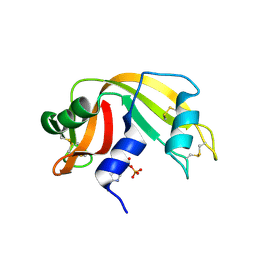 | | RNASE A-NADP COMPLEX | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, RIBONUCLEASE PANCREATIC | | Authors: | Chavali, G.B, Holloway, D.E, Baker, M.D, Acharya, K.R. | | Deposit date: | 2008-12-10 | | Release date: | 2009-02-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Influence of Naturally-Occurring 5'-Pyrophosphate-Linked Substituents on the Binding of Adenylic Inhibitors to Ribonuclease A: An X-Ray Crystallographic Study.
Biopolymers, 91, 2009
|
|
1BL5
 
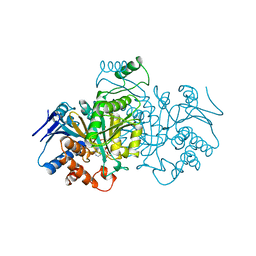 | | ISOCITRATE DEHYDROGENASE FROM E. COLI SINGLE TURNOVER LAUE STRUCTURE OF RATE-LIMITED PRODUCT COMPLEX, 10 MSEC TIME RESOLUTION | | Descriptor: | 2-OXOGLUTARIC ACID, ISOCITRATE DEHYDROGENASE, MAGNESIUM ION, ... | | Authors: | Stoddard, B.L, Cohen, B, Brubaker, M, Mesecar, A, Koshland Junior, D.E. | | Deposit date: | 1998-07-23 | | Release date: | 1999-05-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Millisecond Laue structures of an enzyme-product complex using photocaged substrate analogs.
Nat.Struct.Biol., 5, 1998
|
|
1BOF
 
 | | GI-ALPHA-1 BOUND TO GDP AND MAGNESIUM | | Descriptor: | GI ALPHA 1, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Coleman, D.E, Sprang, S.R. | | Deposit date: | 1998-08-04 | | Release date: | 1999-01-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of the G protein Gi alpha 1 complexed with GDP and Mg2+: a crystallographic titration experiment.
Biochemistry, 37, 1998
|
|
1EEM
 
 | | GLUTATHIONE TRANSFERASE FROM HOMO SAPIENS | | Descriptor: | GLUTATHIONE, GLUTATHIONE-S-TRANSFERASE, SULFATE ION | | Authors: | Board, P, Coggan, M, Chelvanayagam, G, Easteal, S, Jermiin, L.S, Schulte, G.K, Danley, D.E, Hoth, L.R, Griffor, M.C, Kamath, A.V, Rosner, M.H, Chrunyk, B.A, Perregaux, D.E, Gabel, C.A, Geoghegan, K.F, Pandit, J. | | Deposit date: | 2000-02-01 | | Release date: | 2000-08-11 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification, characterization, and crystal structure of the Omega class glutathione transferases.
J.Biol.Chem., 275, 2000
|
|
4LTZ
 
 | | F95M Epi-isozizaene synthase: Complex with Mg, inorganic pyrophosphate and benzyl triethyl ammonium cation | | Descriptor: | Epi-isozizaene synthase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Li, R, Chou, W, Himmelberger, J.A, Litwin, K, Harris, G, Cane, D.E, Christianson, D.W. | | Deposit date: | 2013-07-24 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.448 Å) | | Cite: | Reprogramming the Chemodiversity of Terpenoid Cyclization by Remolding the Active Site Contour of epi-Isozizaene Synthase.
Biochemistry, 53, 2014
|
|
4LUU
 
 | | V329A Epi-isozizaene synthase: Complex with Mg, inorganic pyrophosphate and benzyl triethyl ammonium cation | | Descriptor: | Epi-isozizaene synthase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Li, R, Chou, W, Himmelberger, J.A, Litwin, K, Harris, G, Cane, D.E, Christianson, D.W. | | Deposit date: | 2013-07-25 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Reprogramming the Chemodiversity of Terpenoid Cyclization by Remolding the Active Site Contour of epi-Isozizaene Synthase.
Biochemistry, 53, 2014
|
|
6GW6
 
 | |
4LXW
 
 | | L72V Epi-isozizaene synthase: Complex with Mg, inorganic pyrophosphate and benzyl triethyl ammonium cation | | Descriptor: | Epi-isozizaene synthase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Li, R, Chou, W, Himmelberger, J.A, Litwin, K, Harris, G, Cane, D.E, Christianson, D.W. | | Deposit date: | 2013-07-30 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.092 Å) | | Cite: | Reprogramming the Chemodiversity of Terpenoid Cyclization by Remolding the Active Site Contour of epi-Isozizaene Synthase.
Biochemistry, 53, 2014
|
|
4V41
 
 | | E. COLI (LAC Z) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-MONOCLINIC) | | Descriptor: | BETA-GALACTOSIDASE, MAGNESIUM ION | | Authors: | Juers, D.H, Jacobson, R.H, Wigley, D, Zhang, X.J, Huber, R.E, Tronrud, D.E, Matthews, B.W. | | Deposit date: | 2000-06-07 | | Release date: | 2014-07-09 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | High resolution refinement of beta-galactosidase in a new crystal form reveals multiple metal-binding sites and provides a structural basis for alpha-complementation.
Protein Sci., 9, 2000
|
|
4V4S
 
 | | Crystal structure of the whole ribosomal complex. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Petry, S, Brodersen, D.E, Murphy IV, F.V, Dunham, C.M, Selmer, M, Tarry, M.J, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2005-10-12 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (6.76 Å) | | Cite: | Crystal Structures of the Ribosome in Complex with Release Factors RF1 and RF2 Bound to a Cognate Stop Codon.
Cell(Cambridge,Mass.), 123, 2005
|
|
4V4T
 
 | | Crystal structure of the whole ribosomal complex with a stop codon in the A-site. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Petry, S, Brodersen, D.E, Murphy IV, F.V, Dunham, C.M, Selmer, M, Tarry, M.J, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2005-10-12 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (6.46 Å) | | Cite: | Crystal Structures of the Ribosome in Complex with Release Factors RF1 and RF2 Bound to a Cognate Stop Codon.
Cell(Cambridge,Mass.), 123, 2005
|
|
