6REK
 
 | | Crystal structure of Pizza6-SH with Cu2+ | | Descriptor: | COPPER (II) ION, GLYCEROL, Pizza6-SH | | Authors: | Noguchi, H, Clarke, D.E, Gryspeerdt, J.L, Feyter, S.D, Voet, A.R.D. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Artificial beta-propeller protein-based hydrolases.
Chem.Commun.(Camb.), 55, 2019
|
|
1MRP
 
 | | FERRIC-BINDING PROTEIN FROM HAEMOPHILUS INFLUENZAE | | Descriptor: | FE (III) ION, FERRIC IRON BINDING PROTEIN, PHOSPHATE ION | | Authors: | Bruns, C.M, Nowalk, A.J, Arvai, A.S, Mctigue, M.A, Vaughan, K.G, Mietzner, T.A, Mcree, D.E. | | Deposit date: | 1997-05-14 | | Release date: | 1998-01-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of Haemophilus influenzae Fe(+3)-binding protein reveals convergent evolution within a superfamily.
Nat.Struct.Biol., 4, 1997
|
|
6P0P
 
 | | Human beta-tryptase co-crystal structure with 5-{4-[3-(aminomethyl)phenyl]piperidine-1-carbonyl}-2-(3'-{4-[3-(aminomethyl)phenyl]piperidine-1-carbonyl}-[1,1'-biphenyl]-3-yl)-2-hydroxy-2H-1,3,2-benzodioxaborol-2-uide | | Descriptor: | (3'-{4-[3-(aminomethyl)phenyl]piperidine-1-carbonyl}[1,1'-biphenyl]-3-yl){4-[3-(aminomethyl)phenyl]piperidin-1-yl}[3,4-di(hydroxy-kappaO)phenyl]methanonato(2-)hydroxyborate(1-), 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, SULFATE ION, ... | | Authors: | Giardina, S.F, Pingle, M.R, Werner, D.S, Feinberg, P.B, Foreman, K.W, Bergstrom, D.E, Arnold, L.D, Barany, F. | | Deposit date: | 2019-05-17 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Novel, Self-Assembling Dimeric Inhibitors of Human beta Tryptase.
J.Med.Chem., 63, 2020
|
|
1DI1
 
 | | CRYSTAL STRUCTURE OF ARISTOLOCHENE SYNTHASE FROM PENICILLIUM ROQUEFORTI | | Descriptor: | ARISTOLOCHENE SYNTHASE | | Authors: | Caruthers, J.M, Kang, I, Cane, D.E, Christianson, D.W, Rynkiewicz, M.J. | | Deposit date: | 1999-11-28 | | Release date: | 2000-08-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure determination of aristolochene synthase from the blue cheese mold, Penicillium roqueforti.
J.Biol.Chem., 275, 2000
|
|
6P3O
 
 | | Tetrahydroprotoberberine N-methyltransferase in complex with (S)-cis-N-methylstylopine and S-adenosylhomocysteine | | Descriptor: | (5S,12bS)-5-methyl-6,7,12b,13-tetrahydro-2H,4H,10H-[1,3]dioxolo[4,5-g][1,3]dioxolo[7,8]isoquinolino[3,2-a]isoquinolin-5-ium, S-ADENOSYL-L-HOMOCYSTEINE, Tetrahydroprotoberberine N-methyltransferase | | Authors: | Lang, D.E, Morris, J.S, Rowley, M, Torres, M.A, Maksimovich, V.A, Facchini, P.J, Ng, K.K.S. | | Deposit date: | 2019-05-24 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-function studies of tetrahydroprotoberberineN-methyltransferase reveal the molecular basis of stereoselective substrate recognition.
J.Biol.Chem., 294, 2019
|
|
1D9Y
 
 | | NEISSERIA GONORRHOEAE FERRIC BINDING PROTEIN | | Descriptor: | FE (III) ION, PHOSPHATE ION, PROTEIN (PERIPLASMIC IRON-BINDING PROTEIN) | | Authors: | McRee, D.E, Bruns, C.M, Williams, P.A, Mietzner, T.A, Nunn, R. | | Deposit date: | 1999-10-30 | | Release date: | 1999-11-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis of Iron Uptake in the Pathogen Neisseria gonorrhoeae
To be Published
|
|
6P3M
 
 | | Tetrahydroprotoberberine N-methyltransferase in complex with S-adenosylhomocysteine | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, tetrahydroprotoberberine N-methyltransferase | | Authors: | Lang, D.E, Morris, J.S, Rowley, M, Torres, M.A, Maksimovich, V.A, Facchini, P.J, Ng, K.K.S. | | Deposit date: | 2019-05-24 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-function studies of tetrahydroprotoberberineN-methyltransferase reveal the molecular basis of stereoselective substrate recognition.
J.Biol.Chem., 294, 2019
|
|
6REI
 
 | | Crystal structure of Pizza6-S with Cd2+ | | Descriptor: | CADMIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Noguchi, H, Clarke, D.E, Gryspeerdt, J.L, Feyter, S.D, Voet, A.R.D. | | Deposit date: | 2019-04-12 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Artificial beta-Propeller Protein-based Hydrolases
To Be Published
|
|
6REM
 
 | | Crystal structure of Pizza6-SH with Sulphate ion | | Descriptor: | Pizza6-SH, SULFATE ION | | Authors: | Noguchi, H, Clarke, D.E, Gryspeerdt, J.L, Feyter, S.D, Voet, A.R.D. | | Deposit date: | 2019-04-12 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Artificial beta-Propeller Protein-based Hydrolases
To Be Published
|
|
6REL
 
 | | Crystal structure of Pizza6-SH with CdCl2 nanocrystal | | Descriptor: | CADMIUM ION, CHLORIDE ION, Pizza6-SH | | Authors: | Noguchi, H, Clarke, D.E, Gryspeerdt, J.L, Feyter, S.D, Voet, A.R.D. | | Deposit date: | 2019-04-12 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Artificial beta-Propeller Protein-based Hydrolases
To Be Published
|
|
6P3N
 
 | | Tetrahydroprotoberberine N-methyltransferase in complex with S-adenosylmethionine | | Descriptor: | S-ADENOSYLMETHIONINE, tetrahydroprotoberberine N-methyltransferase | | Authors: | Lang, D.E, Morris, J.S, Rowley, M, Torres, M.A, Maksimovich, V.A, Facchini, P.J, Ng, K.K.S. | | Deposit date: | 2019-05-24 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-function studies of tetrahydroprotoberberineN-methyltransferase reveal the molecular basis of stereoselective substrate recognition.
J.Biol.Chem., 294, 2019
|
|
1N46
 
 | | CRYSTAL STRUCTURE OF HUMAN TR BETA LIGAND-BINDING DOMAIN COMPLEXED WITH A POTENT SUBTYPE-SELECTIVE THYROMIMETIC | | Descriptor: | Thyroid hormone receptor Beta-1, [4-(4-HYDROXY-3-ISOPROPYL-PHENOXY)-3,5-DIMETHYL-PHENYL]-6-AZAURACIL | | Authors: | Dow, R.L, Schneider, S.R, Paight, E.S, Hank, R.F, Chiang, P, Cornelius, P, Lee, E, Newsome, W.P, Swick, A.G, Spitzer, J, Hargrove, D.M, Patterson, T.A, Pandit, J, Chrunyk, B.A, LeMotte, P.K, Danley, D.E, Rosner, M.H, Ammirati, M.J, Simons, S.P, Schulte, G.K, Tate, B.F, DaSilva-Jardine, P. | | Deposit date: | 2002-10-30 | | Release date: | 2003-04-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of a Novel Series of 6-Azauracil-Based Thyroid Hormone Receptor Ligands:
Potent, TRbeta Subtype-Selective Thyromimetics
Bioorg.Med.Chem.Lett., 13, 2003
|
|
1T4S
 
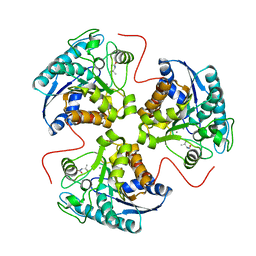 | | arginase-L-valine complex | | Descriptor: | Arginase 1, MANGANESE (II) ION, VALINE | | Authors: | Cama, E, Pethe, S, Boucher, J.-L, Shoufa, H, Emig, F.A, Ash, D.E, Viola, R.E, Mansuy, D, Christianson, D.W. | | Deposit date: | 2004-04-30 | | Release date: | 2004-10-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Inhibitor coordination interactions in the binuclear manganese cluster of arginase
Biochemistry, 43, 2004
|
|
1DB6
 
 | | SOLUTION STRUCTURE OF THE DNA APTAMER 5'-CGACCAACGTGTCGCCTGGTCG-3' COMPLEXED WITH ARGININAMIDE | | Descriptor: | ARGININEAMIDE, DNA | | Authors: | Robertson, S.A, Harada, K, Frankel, A.D, Wemmer, D.E. | | Deposit date: | 1999-11-02 | | Release date: | 2000-02-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure determination and binding kinetics of a DNA aptamer-argininamide complex.
Biochemistry, 39, 2000
|
|
1N8C
 
 | | Solution Structure of a Cis-Opened (10R)-N6-Deoxyadenosine Adduct of (9S,10R)-(9,10)-Epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene in a DNA Duplex | | Descriptor: | (9S,10R)-9-HYDROXY-7,8,9,10-TETRAHYDROBENZO[A]PYRENE, 5'-D(*CP*CP*TP*CP*GP*TP*GP*AP*CP*CP*G)-3', 5'-D(*CP*GP*GP*TP*CP*AP*CP*GP*AP*GP*G)-3' | | Authors: | Volk, D.E, Thiviyanathan, V, Rice, J.S, Luxon, B.A, Shah, J.H, Yagi, H, Sayer, J.M, Yeh, H.J.C, Jerina, D.M, Gorenstein, D.G. | | Deposit date: | 2002-11-20 | | Release date: | 2003-02-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a Cis-Opened (10R)-N6-Deoxyadenosine Adduct of (9S,10R)-(9,10)-Epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene in a DNA Duplex
Biochemistry, 42, 2003
|
|
4QNH
 
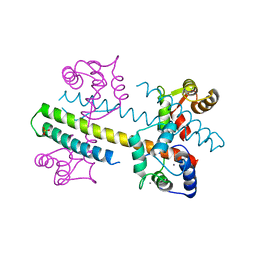 | | Calcium-calmodulin (T79D) complexed with the calmodulin binding domain from a small conductance potassium channel SK2-a | | Descriptor: | CALCIUM ION, Calmodulin, SULFATE ION, ... | | Authors: | Zhang, M, Pascal, J.M, Logothetis, D.E, Zhang, J.F. | | Deposit date: | 2014-06-17 | | Release date: | 2014-08-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Selective phosphorylation modulates the PIP2 sensitivity of the CaM-SK channel complex.
Nat.Chem.Biol., 10, 2014
|
|
1T67
 
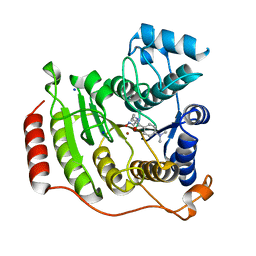 | | Crystal Structure of Human HDAC8 complexed with MS-344 | | Descriptor: | 4-(dimethylamino)-N-[7-(hydroxyamino)-7-oxoheptyl]benzamide, Histone deacetylase 8, SODIUM ION, ... | | Authors: | Somoza, J.R, Skene, R.J, Katz, B.A, Mol, C, Ho, J.D, Jennings, A.J, Luong, C, Arvai, A, Buggy, J.J, Chi, E, Tang, J, Sang, B.-C, Verner, E, Wynands, R, Leahy, E.M, Dougan, D.R, Snell, G, Navre, M, Knuth, M.W, Swanson, R.V, McRee, D.E, Tari, L.W. | | Deposit date: | 2004-05-05 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural Snapshots of Human HDAC8 Provide Insights into the Class I Histone Deacetylases
Structure, 12, 2004
|
|
1T4R
 
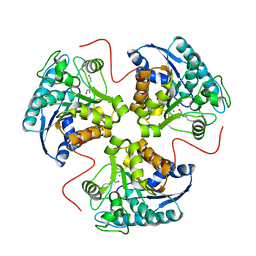 | | arginase-descarboxy-nor-NOHA complex | | Descriptor: | 3-{[(E)-AMINO(HYDROXYIMINO)METHYL]AMINO}PROPAN-1-AMINIUM, Arginase 1, MANGANESE (II) ION | | Authors: | Cama, E, Pethe, S, Boucher, J.-L, Shoufa, H, Emig, F.A, Ash, D.E, Viola, R.E, Mansuy, D, Christianson, D.W. | | Deposit date: | 2004-04-30 | | Release date: | 2005-04-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Inhibitor coordination interactions in the binuclear manganese cluster of arginase
Biochemistry, 43, 2004
|
|
1DGP
 
 | | ARISTOLOCHENE SYNTHASE FARNESOL COMPLEX | | Descriptor: | (2E,6E)-3,7,11-trimethyldodeca-2,6,10-trien-1-ol, (2Z,6Z)-3,7,11-trimethyldodeca-2,6,10-trien-1-ol, ARISTOLOCHENE SYNTHASE | | Authors: | Caruthers, J.M, Kang, I, Cane, D.E, Christianson, D.W. | | Deposit date: | 1999-11-24 | | Release date: | 2001-02-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure determination of aristolochene synthase from the blue cheese mold, Penicillium roqueforti.
J.Biol.Chem., 275, 2000
|
|
1AEE
 
 | | SPECIFICITY OF LIGAND BINDING TO A BURIED POLAR CAVITY AT THE ACTIVE SITE OF CYTOCHROME C PEROXIDASE (ANILINE) | | Descriptor: | ANILINE, CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Musah, R.A, Jensen, G.M, Fitzgerald, M.M, Mcree, D.E, Goodin, D.B. | | Deposit date: | 1997-02-24 | | Release date: | 1997-09-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Artificial protein cavities as specific ligand-binding templates: characterization of an engineered heterocyclic cation-binding site that preserves the evolved specificity of the parent protein.
J.Mol.Biol., 315, 2002
|
|
1TBW
 
 | | Ligand Induced Conformational Shift in the N-terminal Domain of GRP94, Open Conformation | | Descriptor: | ADENOSINE MONOPHOSPHATE, Endoplasmin, MAGNESIUM ION, ... | | Authors: | Gewirth, D.T, Immormino, R.M, Dollins, D.E, Shaffer, P.L, Walker, M.A, Soldano, K.L. | | Deposit date: | 2004-05-20 | | Release date: | 2004-08-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Ligand-induced Conformational Shift in the N-terminal Domain of GRP94, an Hsp90 Chaperone.
J.Biol.Chem., 279, 2004
|
|
1D9V
 
 | |
1DK9
 
 | | SOLUTION STRUCTURE ANALYSIS OF THE DNA DUPLEX D(CATGAGTAC)D(GTACTCATG) | | Descriptor: | 5'-D(CP*AP*TP*GP*AP*GP*TP*AP*CP*)-3', 5'-D(GP*TP*AP*CP*TP*CP*AP*TP*GP*)-3' | | Authors: | Klewer, D.A, Hoskins, A, Davisson, V.J, Bergstrom, D.E, LiWang, A.C. | | Deposit date: | 1999-12-06 | | Release date: | 1999-12-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a DNA duplex containing nucleoside analog 1-(2'-deoxy-beta-D-ribofuranosyl)-3-nitropyrrole and the structure of the unmodified control.
Nucleic Acids Res., 28, 2000
|
|
1DF8
 
 | | S45A MUTANT OF STREPTAVIDIN IN COMPLEX WITH BIOTIN | | Descriptor: | BIOTIN, PROTEIN (STREPTAVIDIN) | | Authors: | Hyre, D.E, Le Trong, I, Freitag, S, Stenkamp, R.E, Stayton, P.S. | | Deposit date: | 1999-11-18 | | Release date: | 2000-09-20 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Ser45 plays an important role in managing both the equilibrium and transition state energetics of the streptavidin-biotin system.
Protein Sci., 9, 2000
|
|
1OJC
 
 | |
